Structural insight into the unique substrate binding mechanism and flavin redox state of UDP-galactopyranose mutase from Aspergillus fumigatus
- PMID: 22334662
- PMCID: PMC3322874
- DOI: 10.1074/jbc.M111.322974
Structural insight into the unique substrate binding mechanism and flavin redox state of UDP-galactopyranose mutase from Aspergillus fumigatus
Abstract
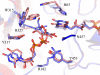
UDP-galactopyranose mutase (UGM) is a flavin-containing enzyme that catalyzes the reversible conversion of UDP-galactopyranose (UDP-Galp) to UDP-galactofuranose (UDP-Galf). As in prokaryotic UGMs, the flavin needs to be reduced for the enzyme to be active. Here we present the first eukaryotic UGM structures from Aspergillus fumigatus (AfUGM). The structures are of UGM alone, with the substrate UDP-Galp and with the inhibitor UDP. Additionally, we report the structures of AfUGM bound to substrate with oxidized and reduced flavin. These structures provide insight into substrate recognition and structural changes observed upon substrate binding involving the mobile loops and the critical arginine residues Arg-182 and Arg-327. Comparison with prokaryotic UGM reveals that despite low sequence identity with known prokaryotic UGMs the overall fold is largely conserved. Structural differences between prokaryotic UGM and AfUGM result from inserts in AfUGM. A notable difference from prokaryotic UGMs is that AfUGM contains a third flexible loop (loop III) above the si-face of the isoalloxazine ring that changes position depending on the redox state of the flavin cofactor. This loop flipping has not been observed in prokaryotic UGMs. In addition we have determined the crystals structures and steady-state kinetic constants of the reaction catalyzed by mutants R182K, R327K, R182A, and R327A. These results support our hypothesis that Arg-182 and Arg-327 play important roles in stabilizing the position of the diphosphates of the nucleotide sugar and help to facilitate the positioning of the galactose moiety for catalysis.
Figures


Similar articles
-
Targeting UDP-galactopyranose mutases from eukaryotic human pathogens.Curr Pharm Des. 2013;19(14):2561-73. doi: 10.2174/1381612811319140007. Curr Pharm Des. 2013. PMID: 23116395 Free PMC article. Review.
-
Crystal structures and small-angle x-ray scattering analysis of UDP-galactopyranose mutase from the pathogenic fungus Aspergillus fumigatus.J Biol Chem. 2012 Mar 16;287(12):9041-51. doi: 10.1074/jbc.M111.327536. Epub 2012 Jan 31. J Biol Chem. 2012. PMID: 22294687 Free PMC article.
-
Contributions of unique active site residues of eukaryotic UDP-galactopyranose mutases to substrate recognition and active site dynamics.Biochemistry. 2014 Dec 16;53(49):7794-804. doi: 10.1021/bi501008z. Epub 2014 Dec 2. Biochemistry. 2014. PMID: 25412209 Free PMC article.
-
Characterization of recombinant UDP-galactopyranose mutase from Aspergillus fumigatus.Arch Biochem Biophys. 2010 Oct 1;502(1):31-8. doi: 10.1016/j.abb.2010.06.035. Epub 2010 Jul 6. Arch Biochem Biophys. 2010. PMID: 20615386
-
Structure, mechanism, and dynamics of UDP-galactopyranose mutase.Arch Biochem Biophys. 2014 Feb 15;544:128-41. doi: 10.1016/j.abb.2013.09.017. Epub 2013 Oct 3. Arch Biochem Biophys. 2014. PMID: 24096172 Free PMC article. Review.
Cited by
-
Aspergillus nidulans cell wall composition and function change in response to hosting several Aspergillus fumigatus UDP-galactopyranose mutase activity mutants.PLoS One. 2014 Jan 15;9(1):e85735. doi: 10.1371/journal.pone.0085735. eCollection 2014. PLoS One. 2014. PMID: 24454924 Free PMC article.
-
Conformational Control of UDP-Galactopyranose Mutase Inhibition.Biochemistry. 2017 Aug 1;56(30):3983-3992. doi: 10.1021/acs.biochem.7b00189. Epub 2017 Jul 20. Biochemistry. 2017. PMID: 28608671 Free PMC article.
-
Targeting UDP-galactopyranose mutases from eukaryotic human pathogens.Curr Pharm Des. 2013;19(14):2561-73. doi: 10.2174/1381612811319140007. Curr Pharm Des. 2013. PMID: 23116395 Free PMC article. Review.
-
UDP-galactopyranose mutase in nematodes.Biochemistry. 2013 Jun 25;52(25):4391-8. doi: 10.1021/bi400264d. Epub 2013 Jun 11. Biochemistry. 2013. PMID: 23697711 Free PMC article.
-
UDP-galactopyranose mutases from Leishmania species that cause visceral and cutaneous leishmaniasis.Arch Biochem Biophys. 2013 Oct 15;538(2):103-10. doi: 10.1016/j.abb.2013.08.014. Epub 2013 Sep 3. Arch Biochem Biophys. 2013. PMID: 24012809 Free PMC article.
References
-
- Tefsen B., Ram A. F., Van Die I., Routier F. H. (2012) Galactofuranose in eukaryotes. Aspects of biosynthesis and functional impact. Glycobiology 22, 456–469 - PubMed
-
- Richards M. R., Lowary T. L. (2009) Chemistry and biology of galactofuranose-containing polysaccharides. ChemBioChem 10, 1920–1938 - PubMed
-
- Chad J. M., Sarathy K. P., Gruber T. D., Addala E., Kiessling L. L., Sanders D. A. (2007) Site-directed mutagenesis of UDP-galactopyranose mutase reveals a critical role for the active-site, conserved arginine residues. Biochemistry 46, 6723–6732 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

