Kindlin-3 mediates integrin αLβ2 outside-in signaling, and it interacts with scaffold protein receptor for activated-C kinase 1 (RACK1)
- PMID: 22334666
- PMCID: PMC3322817
- DOI: 10.1074/jbc.M111.299594
Kindlin-3 mediates integrin αLβ2 outside-in signaling, and it interacts with scaffold protein receptor for activated-C kinase 1 (RACK1)
Abstract
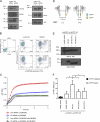
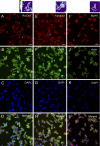
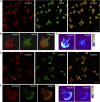
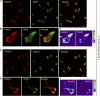
Integrins are heterodimeric type I membrane cell adhesion molecules that are involved in many biological processes. Integrins are bidirectional signal transducers because their cytoplasmic tails are docking sites for cytoskeletal and signaling molecules. Kindlins are cytoplasmic molecules that mediate inside-out signaling and activation of the integrins. The three kindlin paralogs in humans are kindlin-1, -2, and -3. Each of these contains a 4.1-ezrin-radixin-moesin (FERM) domain and a pleckstrin homology domain. Kindlin-3 is expressed in platelets, hematopoietic cells, and endothelial cells. Here we show that kindlin-3 is involved in integrin αLβ2 outside-in signaling. It also promotes micro-clustering of integrin αLβ2. We provide evidence that kindlin-3 interacts with the receptor for activated-C kinase 1 (RACK1), a scaffold protein that folds into a seven-blade propeller. This interaction involves the pleckstrin homology domain of kindlin-3 and blades 5-7 of RACK1. Using the SKW3 human T lymphoma cells, we show that integrin αLβ2 engagement by its ligand ICAM-1 promotes the association of kindlin-3 with RACK1. We also show that kindlin-3 co-localizes with RACK1 in polarized SKW3 cells and human T lymphoblasts. Our findings suggest that kindlin-3 plays an important role in integrin αLβ2 outside-in signaling.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

