Role of N-glycosylation sites and CXC motifs in trafficking of medicago truncatula Nod factor perception protein to plasma membrane
- PMID: 22334694
- PMCID: PMC3322848
- DOI: 10.1074/jbc.M111.281634
Role of N-glycosylation sites and CXC motifs in trafficking of medicago truncatula Nod factor perception protein to plasma membrane
Abstract
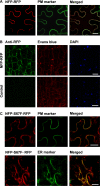
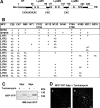
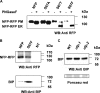
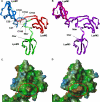
The lysin motif receptor-like kinase, NFP (Nod factor perception), is a key protein in the legume Medicago truncatula for the perception of lipochitooligosaccharidic Nod factors, which are secreted bacterial signals essential for establishing the nitrogen-fixing legume-rhizobia symbiosis. Predicted structural and genetic analyses strongly suggest that NFP is at least part of a Nod factor receptor, but few data are available about this protein. Characterization of a variant encoded by the mutant allele nfp-2 revealed the sensitivity of this protein to the endoplasmic reticulum quality control mechanisms, affecting its trafficking to the plasma membrane. Further analysis revealed that the extensive N-glycosylation of the protein is not essential for biological activity. In the NFP extracellular region, two CXC motifs and two other Cys residues were found to be involved in disulfide bridges, and these are necessary for correct folding and localization of the protein. Analysis of the intracellular region revealed its importance for biological activity but suggests that it does not rely on kinase activity. This work shows that NFP trafficking to the plasma membrane is highly sensitive to regulation in the endoplasmic reticulum and has identified structural features of the protein, particularly disulfide bridges involving CXC motifs in the extracellular region that are required for its biological function.
Figures

Similar articles
-
LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors.Glycobiology. 2006 Sep;16(9):801-9. doi: 10.1093/glycob/cwl006. Epub 2006 May 24. Glycobiology. 2006. PMID: 16723404
-
The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes.Plant Physiol. 2006 Sep;142(1):265-79. doi: 10.1104/pp.106.084657. Epub 2006 Jul 14. Plant Physiol. 2006. PMID: 16844829 Free PMC article.
-
Nod factor perception protein carries weight in biotic interactions.Trends Plant Sci. 2013 Oct;18(10):566-74. doi: 10.1016/j.tplants.2013.06.001. Epub 2013 Jul 10. Trends Plant Sci. 2013. PMID: 23850222 Review.
-
Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis.PLoS One. 2011;6(11):e26114. doi: 10.1371/journal.pone.0026114. Epub 2011 Nov 8. PLoS One. 2011. PMID: 22087221 Free PMC article.
-
Annexins - calcium- and membrane-binding proteins in the plant kingdom: potential role in nodulation and mycorrhization in Medicago truncatula.Acta Biochim Pol. 2009;56(2):199-210. Epub 2009 May 7. Acta Biochim Pol. 2009. PMID: 19421430 Review.
Cited by
-
LysM Receptor-Like Kinase and LysM Receptor-Like Protein Families: An Update on Phylogeny and Functional Characterization.Front Plant Sci. 2018 Oct 24;9:1531. doi: 10.3389/fpls.2018.01531. eCollection 2018. Front Plant Sci. 2018. PMID: 30405668 Free PMC article. Review.
-
Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity.Plant Cell. 2012 Aug;24(8):3406-19. doi: 10.1105/tpc.112.102475. Epub 2012 Aug 7. Plant Cell. 2012. PMID: 22872757 Free PMC article.
-
di-Cysteine motifs in the C-terminus of plant HMA4 proteins confer nanomolar affinity for zinc and are essential for HMA4 function in vivo.J Exp Bot. 2018 Nov 26;69(22):5547-5560. doi: 10.1093/jxb/ery311. J Exp Bot. 2018. PMID: 30137564 Free PMC article.
-
PUB1 Interacts with the Receptor Kinase DMI2 and Negatively Regulates Rhizobial and Arbuscular Mycorrhizal Symbioses through Its Ubiquitination Activity in Medicago truncatula.Plant Physiol. 2016 Apr;170(4):2312-24. doi: 10.1104/pp.15.01694. Epub 2016 Feb 2. Plant Physiol. 2016. PMID: 26839127 Free PMC article.
-
The receptor-like cytoplasmic kinase AeRLCK2 mediates Nod-independent rhizobial symbiosis in Aeschynomene legumes.Plant Cell. 2025 Aug 4;37(8):koaf201. doi: 10.1093/plcell/koaf201. Plant Cell. 2025. PMID: 40811611 Free PMC article.
References
-
- Gough C., Cullimore J. (2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant Microbe Interact. 24, 867–878 - PubMed
-
- Oldroyd G. E., Downie J. A. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546 - PubMed
-
- Arrighi J. F., Barre A., Ben Amor B., Bersoult A., Soriano L. C., Mirabella R., de Carvalho-Niebel F., Journet E. P., Ghérardi M., Huguet T., Geurts R., Dénarié J., Rougé P., Gough C. (2006) The Medicago truncatula lysin [corrected] motif receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279 - PMC - PubMed
-
- Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640 - PubMed
-
- Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

