Single-stranded genomic architecture constrains optimal codon usage
- PMID: 22334868
- PMCID: PMC3278643
- DOI: 10.4161/bact.1.4.18496
Single-stranded genomic architecture constrains optimal codon usage
Abstract
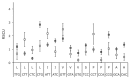
Viral codon usage is shaped by the conflicting forces of mutational pressure and selection to match host patterns for optimal expression. We examined whether genomic architecture (single- or double-stranded DNA) influences the degree to which bacteriophage codon usage differ from their primary bacterial hosts and each other. While both correlated equally with their hosts' genomic nucleotide content, the coat genes of ssDNA phages were less well adapted than those of dsDNA phages to their hosts' codon usage profiles due to their preference for codons ending in thymine. No specific biases were detected in dsDNA phage genomes. In all nine of ten cases of codon redundancy in which a specific codon was overrepresented, ssDNA phages favored the NNT codon. A cytosine to thymine biased mutational pressure working in conjunction with strong selection against non-synonymous mutations appears be shaping codon usage bias in ssDNA viral genomes.
Figures




References
-
- Sharp PM. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986;3:75–83. - PubMed
-
- Shields DC, Sharp PM, Higgins DG, Wright F. “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988;5:704–16. - PubMed
LinkOut - more resources
Full Text Sources
