Truncations of titin causing dilated cardiomyopathy
- PMID: 22335739
- PMCID: PMC3660031
- DOI: 10.1056/NEJMoa1110186
Truncations of titin causing dilated cardiomyopathy
Abstract
Background: Dilated cardiomyopathy and hypertrophic cardiomyopathy arise from mutations in many genes. TTN, the gene encoding the sarcomere protein titin, has been insufficiently analyzed for cardiomyopathy mutations because of its enormous size.
Methods: We analyzed TTN in 312 subjects with dilated cardiomyopathy, 231 subjects with hypertrophic cardiomyopathy, and 249 controls by using next-generation or dideoxy sequencing. We evaluated deleterious variants for cosegregation in families and assessed clinical characteristics.
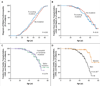
Results: We identified 72 unique mutations (25 nonsense, 23 frameshift, 23 splicing, and 1 large tandem insertion) that altered full-length titin. Among subjects studied by means of next-generation sequencing, the frequency of TTN mutations was significantly higher among subjects with dilated cardiomyopathy (54 of 203 [27%]) than among subjects with hypertrophic cardiomyopathy (3 of 231 [1%], P=3×10(-16)) or controls (7 of 249 [3%], P=9×10(-14)). TTN mutations cosegregated with dilated cardiomyopathy in families (combined lod score, 11.1) with high (>95%) observed penetrance after the age of 40 years. Mutations associated with dilated cardiomyopathy were overrepresented in the titin A-band but were absent from the Z-disk and M-band regions of titin (P≤0.01 for all comparisons). Overall, the rates of cardiac outcomes were similar in subjects with and those without TTN mutations, but adverse events occurred earlier in male mutation carriers than in female carriers (P=4×10(-5)).
Conclusions: TTN truncating mutations are a common cause of dilated cardiomyopathy, occurring in approximately 25% of familial cases of idiopathic dilated cardiomyopathy and in 18% of sporadic cases. Incorporation of sequencing approaches that detect TTN truncations into genetic testing for dilated cardiomyopathy should substantially increase test sensitivity, thereby allowing earlier diagnosis and therapeutic intervention for many patients with dilated cardiomyopathy. Defining the functional effects of TTN truncating mutations should improve our understanding of the pathophysiology of dilated cardiomyopathy. (Funded by the Howard Hughes Medical Institute and others.).
Figures

Comment in
-
Cardiomyopathies: Mutations in a big gene are linked to a big heart.Nat Rev Cardiol. 2012 Mar 6;9(4):185. doi: 10.1038/nrcardio.2012.30. Nat Rev Cardiol. 2012. PMID: 22392061 No abstract available.
-
Mutations in the sensitive giant titin result in a broken heart.Circ Res. 2012 Jul 6;111(2):158-61. doi: 10.1161/RES.0b013e3182635ca2. Circ Res. 2012. PMID: 22773424 No abstract available.
References
-
- Olivotto I, Girolami F, Ackerman MJ, et al. Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2008;83:630–638. - PubMed
-
- Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. - PubMed
-
- Michels VV, Moll PP, Miller FA, et al. The frequency of familial dilated cardio-myopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 1992;326:77–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U120085815/MRC_/Medical Research Council/United Kingdom
- U01 HL066582/HL/NHLBI NIH HHS/United States
- N01-HV-48194/HV/NHLBI NIH HHS/United States
- N01 HV048194/HL/NHLBI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 HL080494/HL/NHLBI NIH HHS/United States
- R01 HL084553/HL/NHLBI NIH HHS/United States
- P01 HL059139/HL/NHLBI NIH HHS/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
- U01 HL098166/HL/NHLBI NIH HHS/United States
- SP/10/10/28431/BHF_/British Heart Foundation/United Kingdom
- P50 HL052320/HL/NHLBI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
