TS-AMIR: a topology string alignment method for intensive rapid protein structure comparison
- PMID: 22336468
- PMCID: PMC3298807
- DOI: 10.1186/1748-7188-7-4
TS-AMIR: a topology string alignment method for intensive rapid protein structure comparison
Abstract
Background: In structural biology, similarity analysis of protein structure is a crucial step in studying the relationship between proteins. Despite the considerable number of techniques that have been explored within the past two decades, the development of new alternative methods is still an active research area due to the need for high performance tools.
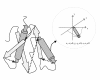
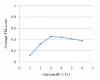
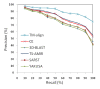
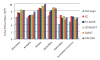
Results: In this paper, we present TS-AMIR, a Topology String Alignment Method for Intensive Rapid comparison of protein structures. The proposed method works in two stages: In the first stage, the method generates a topology string based on the geometric details of secondary structure elements, and then, utilizes an n-gram modelling technique over entropy concept to capture similarities in these strings. This initial correspondence map between secondary structure elements is submitted to the second stage in order to obtain the alignment at the residue level. Applying the Kabsch method, a heuristic step-by-step algorithm is adopted in the second stage to align the residues, resulting in an optimal rotation matrix and minimized RMSD. The performance of the method was assessed in different information retrieval tests and the results were compared with those of CE and TM-align, representing two geometrical tools, and YAKUSA, 3D-BLAST and SARST as three representatives of linear encoding schemes. It is shown that the method obtains a high running speed similar to that of the linear encoding schemes. In addition, the method runs about 800 and 7200 times faster than TM-align and CE respectively, while maintaining a competitive accuracy with TM-align and CE.
Conclusions: The experimental results demonstrate that linear encoding techniques are capable of reaching the same high degree of accuracy as that achieved by geometrical methods, while generally running hundreds of times faster than conventional programs.
Figures




References
-
- Gibrat JF, Madej T, Spouge JL, Bryant SH. The VAST protein structure comparison method. Biophysics Jounnal. 1997;72:MP298.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

