Site-specific transgenesis in Xenopus
- PMID: 22337567
- PMCID: PMC3294184
- DOI: 10.1002/dvg.22006
Site-specific transgenesis in Xenopus
Abstract
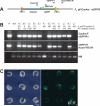
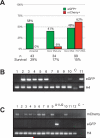
Transgenesis is an essential, powerful tool for investigating gene function and the activities of enhancers, promoters, and transcription factors in the chromatin environment. In Xenopus, current methods generate germ-line transgenics by random insertion, often resulting in mosaicism, position-dependent variations in expression, and lab-to-lab differences in efficiency. We have developed and tested a Xenopus FLP-FRT recombinase-mediated transgenesis (X-FRMT) method. We demonstrate transgenesis of Xenopus laevis by FLP-catalyzed recombination of donor plasmid cassettes into F(1) tadpoles with host cassette transgenes. X-FRMT provides a new method for generating transgenic Xenopus. Once Xenopus lines harboring single host cassettes are generated, X-FRMT should allow for the targeting of transgenes to well-characterized integration site(s), requiring no more special reagents or training than that already common to most Xenopus labs.
Copyright © 2012 Wiley Periodicals, Inc.
Figures


Similar articles
-
CRISPR/Cas9-based simple transgenesis in Xenopus laevis.Dev Biol. 2022 Sep;489:76-83. doi: 10.1016/j.ydbio.2022.06.001. Epub 2022 Jun 9. Dev Biol. 2022. PMID: 35690103
-
A Plasmid Set for Efficient Bacterial Artificial Chromosome (BAC) Transgenesis in Zebrafish.G3 (Bethesda). 2016 Apr 7;6(4):829-34. doi: 10.1534/g3.115.026344. G3 (Bethesda). 2016. PMID: 26818072 Free PMC article.
-
Targeted transgene integration overcomes variability of position effects in zebrafish.Development. 2014 Feb;141(3):715-24. doi: 10.1242/dev.100347. Development. 2014. PMID: 24449846 Free PMC article.
-
PITT: pronuclear injection-based targeted transgenesis, a reliable transgene expression method in mice.Exp Anim. 2012;61(5):489-502. doi: 10.1538/expanim.61.489. Exp Anim. 2012. PMID: 23095812 Review.
-
Site-specific transgene insertion: an approach.Reprod Fertil Dev. 1994;6(5):585-8. doi: 10.1071/rd9940585. Reprod Fertil Dev. 1994. PMID: 7569037 Review.
Cited by
-
Zebrafish transgenic constructs label specific neurons in Xenopus laevis spinal cord and identify frog V0v spinal neurons.Dev Neurobiol. 2017 Sep;77(8):1007-1020. doi: 10.1002/dneu.22490. Epub 2017 Mar 8. Dev Neurobiol. 2017. PMID: 28188691 Free PMC article.
-
Distinct cis-acting regions control six6 expression during eye field and optic cup stages of eye formation.Dev Biol. 2017 Jun 15;426(2):418-428. doi: 10.1016/j.ydbio.2017.04.003. Epub 2017 Apr 21. Dev Biol. 2017. PMID: 28438336 Free PMC article.
-
FLP recombinase-mediated site-specific recombination in silkworm, Bombyx mori.PLoS One. 2012;7(6):e40150. doi: 10.1371/journal.pone.0040150. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768245 Free PMC article.
-
Using transgenic reporters to visualize bone and cartilage signaling during development in vivo.Front Endocrinol (Lausanne). 2012 Jul 18;3:91. doi: 10.3389/fendo.2012.00091. eCollection 2012. Front Endocrinol (Lausanne). 2012. PMID: 22826703 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

