Constant pH Molecular Dynamics Simulations of Nucleic Acids in Explicit Solvent
- PMID: 22337595
- PMCID: PMC3277849
- DOI: 10.1021/ct2006314
Constant pH Molecular Dynamics Simulations of Nucleic Acids in Explicit Solvent
Abstract
The nucleosides of adenine and cytosine have pKa values of 3.50 and 4.08, respectively, and are assumed to be unprotonated under physiological conditions. However, evidence from recent NMR and X-Ray crystallography studies has revealed the prevalence of protonated adenine and cytosine in RNA macromolecules. Such nucleotides with elevated pKa values may play a role in stabilizing RNA structure and participate in the mechanism of ribozyme catalysis. With the work presented here, we establish the framework and demonstrate the first constant pH MD simulations (CPHMD) for nucleic acids in explicit solvent in which the protonation state is coupled to the dynamical evolution of the RNA system via λ-dynamics. We adopt the new functional form λ(Nexp) for λ that was recently developed for Multi-Site λ-Dynamics (MSλD) and demonstrate good sampling characteristics in which rapid and frequent transitions between the protonated and unprotonated states at pH = pKa are achieved. Our calculated pKa values of simple nucleotides are in a good agreement with experimentally measured values, with a mean absolute error of 0.24 pKa units. This work demonstrates that CPHMD can be used as a powerful tool to investigate pH-dependent biological properties of RNA macromolecules.
Figures
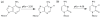
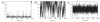
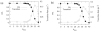


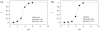

Similar articles
-
Constant pH molecular dynamics of proteins in explicit solvent with proton tautomerism.Proteins. 2014 Jul;82(7):1319-31. doi: 10.1002/prot.24499. Epub 2014 Jan 15. Proteins. 2014. PMID: 24375620 Free PMC article.
-
pH-dependent dynamics of complex RNA macromolecules.J Chem Theory Comput. 2013 Feb 12;9(2):935-943. doi: 10.1021/ct300942z. Epub 2013 Jan 3. J Chem Theory Comput. 2013. PMID: 23525495 Free PMC article.
-
Towards Accurate Prediction of Protonation Equilibrium of Nucleic Acids.J Phys Chem Lett. 2013 Mar 7;4(5):760-766. doi: 10.1021/jz400078d. Epub 2013 Feb 12. J Phys Chem Lett. 2013. PMID: 23526474 Free PMC article.
-
Constant pH Replica Exchange Molecular Dynamics in Explicit Solvent Using Discrete Protonation States: Implementation, Testing, and Validation.J Chem Theory Comput. 2014 Mar 11;10(3):1341-1352. doi: 10.1021/ct401042b. Epub 2014 Feb 5. J Chem Theory Comput. 2014. PMID: 24803862 Free PMC article.
-
Molecular dynamic simulations of environment and sequence dependent DNA conformations: the development of the BMS nucleic acid force field and comparison with experimental results.J Biomol Struct Dyn. 1998 Dec;16(3):487-509. doi: 10.1080/07391102.1998.10508265. J Biomol Struct Dyn. 1998. PMID: 10052609 Review.
Cited by
-
Characterizing the protonation state of cytosine in transient G·C Hoogsteen base pairs in duplex DNA.J Am Chem Soc. 2013 May 8;135(18):6766-9. doi: 10.1021/ja400994e. Epub 2013 Apr 29. J Am Chem Soc. 2013. PMID: 23506098 Free PMC article.
-
Constant pH molecular dynamics of proteins in explicit solvent with proton tautomerism.Proteins. 2014 Jul;82(7):1319-31. doi: 10.1002/prot.24499. Epub 2014 Jan 15. Proteins. 2014. PMID: 24375620 Free PMC article.
-
Constant-pH Simulations with the Polarizable Atomic Multipole AMOEBA Force Field.J Chem Theory Comput. 2024 Apr 9;20(7):2921-2933. doi: 10.1021/acs.jctc.3c01180. Epub 2024 Mar 20. J Chem Theory Comput. 2024. PMID: 38507252 Free PMC article.
-
Electromagnetic Field Stimulation Therapy for Alzheimer's Disease.Neurology (Chic). 2024;3(1):1020. Epub 2024 Jan 5. Neurology (Chic). 2024. PMID: 38699565 Free PMC article.
-
Force Field Limitations of All-Atom Continuous Constant pH Molecular Dynamics.J Phys Chem B. 2024 Nov 28;128(47):11616-11624. doi: 10.1021/acs.jpcb.4c05971. Epub 2024 Nov 12. J Phys Chem B. 2024. PMID: 39531617 Free PMC article.
References
-
- Izatt RM, Christensen JJ, Rytting JH. Chem. Rev. 1971;71:439. - PubMed
-
- Gao XL, Patel DJ. J. Biol. Chem. 1987;262:16973. - PubMed
-
- Asensio JL, Lane AN, Dhesi J, Bergqvist S, Brown T. J. Mol. Biol. 1998;275:811. - PubMed
-
- Jang SB, Hung LW, Chi YI, Holbrook EL, Carter RJ, Holbrook SR. Biochemistry. 1998;37:11726. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
