Kinetics and fidelity of polymerization by DNA polymerase III from Sulfolobus solfataricus
- PMID: 22339170
- PMCID: PMC3624615
- DOI: 10.1021/bi201799a
Kinetics and fidelity of polymerization by DNA polymerase III from Sulfolobus solfataricus
Abstract
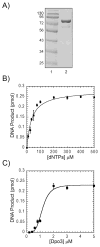
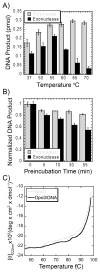
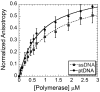
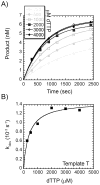
We have biochemically and kinetically characterized the polymerase and exonuclease activities of the third B-family polymerase (Dpo3) from the hyperthermophilic Crenarchaeon, Sulfolobus solfataricus (Sso). We have established through mutagenesis that despite incomplete sequence conservation, the polymerase and exonuclease active sites are functionally conserved in Dpo3. Using pre-steady-state kinetics, we can measure the fidelity of nucleotide incorporation by Dpo3 from the polymerase active site alone to be 10(3)-10(4) at 37 °C. The functional exonuclease proofreading active site will increase fidelity by at least 10(2), making Dpo3 comparable to other DNA polymerases in this family. Additionally, Dpo3's exonuclease activity is modulated by temperature, where a loss of promiscuous degradation activity can be attributed to a reorganization of the exonuclease domain when it is bound to primer-template DNA at high temperatures. Unexpectedly, the DNA binding affinity is weak compared with those of other DNA polymerases of this family. A comparison of the fidelity, polymerization kinetics, and associated functional exonuclease domain with those previously reported for other Sso polymerases (Dpo1 and Dpo4) illustrates that Dpo3 is a potential player in the proper maintenance of the archaeal genome.
Figures


References
-
- Filee J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol. 2002;54:763–773. - PubMed
-
- Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. Eukaryotic DNA polymerases: Proposal for a revised nomenclature. J Biol Chem. 2001;276:43487–43490. - PubMed
-
- Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. - PubMed
-
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. - PubMed
-
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

