Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation
- PMID: 22341441
- PMCID: PMC3479156
- DOI: 10.1016/j.cell.2012.01.018
Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation
Abstract
Understanding the pathway and kinetic mechanisms of transcription initiation is essential for quantitative understanding of gene regulation, but initiation is a multistep process, the features of which can be obscured in bulk analysis. We used a multiwavelength single-molecule fluorescence colocalization approach, CoSMoS, to define the initiation pathway at an activator-dependent bacterial σ(54) promoter that recapitulates characteristic features of eukaryotic promoters activated by enhancer binding proteins. The experiments kinetically characterize all major steps of the initiation process, revealing heretofore unknown features, including reversible formation of two closed complexes with greatly differing stabilities, multiple attempts for each successful formation of an open complex, and efficient release of σ(54) from the polymerase core at the start of transcript synthesis. Open complexes are committed to transcription, suggesting that regulation likely targets earlier steps in the mechanism. CoSMoS is a powerful, generally applicable method to elucidate the mechanisms of transcription and other multistep biochemical processes.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

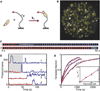



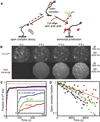

Comment in
-
CoSMoS unravels mysteries of transcription initiation.Cell. 2012 Feb 17;148(4):635-7. doi: 10.1016/j.cell.2012.01.042. Cell. 2012. PMID: 22341438 Free PMC article.
Similar articles
-
Initial events in bacterial transcription initiation.Biomolecules. 2015 May 27;5(2):1035-62. doi: 10.3390/biom5021035. Biomolecules. 2015. PMID: 26023916 Free PMC article. Review.
-
Bacterial RNA polymerase can retain σ70 throughout transcription.Proc Natl Acad Sci U S A. 2016 Jan 19;113(3):602-7. doi: 10.1073/pnas.1513899113. Epub 2016 Jan 5. Proc Natl Acad Sci U S A. 2016. PMID: 26733675 Free PMC article.
-
The bacterial enhancer-binding protein NTRC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation.Genes Dev. 1995 Aug 15;9(16):2042-52. doi: 10.1101/gad.9.16.2042. Genes Dev. 1995. PMID: 7649482
-
Specific binding of the transcription factor sigma-54 to promoter DNA.Nature. 1992 Jul 30;358(6385):422-4. doi: 10.1038/358422a0. Nature. 1992. PMID: 1641025
-
RNA polymerase: in search of promoters.Ann N Y Acad Sci. 2013 Jul;1293:25-32. doi: 10.1111/nyas.12197. Epub 2013 Jul 15. Ann N Y Acad Sci. 2013. PMID: 23855603 Free PMC article. Review.
Cited by
-
A Single-Molecule Surface-Based Platform to Detect the Assembly and Function of the Human RNA Polymerase II Transcription Machinery.Structure. 2020 Dec 1;28(12):1337-1343.e4. doi: 10.1016/j.str.2020.07.009. Epub 2020 Aug 6. Structure. 2020. PMID: 32763141 Free PMC article.
-
Specific anchoring of large topologically closed DNA for single-molecule protein:DNA interactions.Biophys Rep (N Y). 2024 Jan 26;4(1):100144. doi: 10.1016/j.bpr.2024.100144. eCollection 2024 Mar 13. Biophys Rep (N Y). 2024. PMID: 38390466 Free PMC article.
-
Transcriptional activators in the early Drosophila embryo perform different kinetic roles.Cell Syst. 2023 Apr 19;14(4):258-272.e4. doi: 10.1016/j.cels.2023.03.006. Cell Syst. 2023. PMID: 37080162 Free PMC article.
-
Initial events in bacterial transcription initiation.Biomolecules. 2015 May 27;5(2):1035-62. doi: 10.3390/biom5021035. Biomolecules. 2015. PMID: 26023916 Free PMC article. Review.
-
Single-cell measurement of plasmid copy number and promoter activity.Nat Commun. 2021 Mar 5;12(1):1475. doi: 10.1038/s41467-021-21734-y. Nat Commun. 2021. PMID: 33674569 Free PMC article.
References
-
- Bar-Nahum G, Nudler E. Isolation and characterization of σ70-retaining transcription elongation complexes from Escherichia coli. Cell. 2001;106:443–451. - PubMed
-
- Bernardo LM, Johansson LU, Solera D, Skärfstad E, Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of σ54-dependent transcription. Mol. Microbiol. 2006;60:749–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

