Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness
- PMID: 22341444
- PMCID: PMC3285425
- DOI: 10.1016/j.cell.2011.12.027
Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness
Abstract
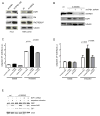
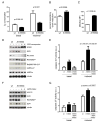
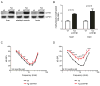
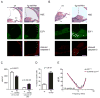
Mitochondrial dysfunction causes poorly understood tissue-specific pathology stemming from primary defects in respiration, coupled with altered reactive oxygen species (ROS), metabolic signaling, and apoptosis. The A1555G mtDNA mutation that causes maternally inherited deafness disrupts mitochondrial ribosome function, in part, via increased methylation of the mitochondrial 12S rRNA by the methyltransferase mtTFB1. In patient-derived A1555G cells, we show that 12S rRNA hypermethylation causes ROS-dependent activation of AMP kinase and the proapoptotic nuclear transcription factor E2F1. This retrograde mitochondrial-stress relay is operative in vivo, as transgenic-mtTFB1 mice exhibit enhanced 12S rRNA methylation in multiple tissues, increased E2F1 and apoptosis in the stria vascularis and spiral ganglion neurons of the inner ear, and progressive E2F1-dependent hearing loss. This mouse mitochondrial disease model provides a robust platform for deciphering the complex tissue specificity of human mitochondrial-based disorders, as well as the precise pathogenic mechanism of maternally inherited deafness and its exacerbation by environmental factors.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

References
-
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. - PubMed
-
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. - PubMed
-
- Bykhovskaya Y, Mengesha E, Wang D, Yang H, Estivill X, Shohat M, Fischel-Ghodsian N. Human mitochondrial transcription factor B1 as a modifier gene for hearing loss associated with the mitochondrial A1555G mutation. Mol Genet Metab. 2004;82:27–32. - PubMed
-
- Chen P, Zindy F, Abdala C, Liu F, Li X, Roussel MF, Segil N. Progressive hearing loss in mice lacking the cyclin-dependent kinase inhibitor Ink4d. Nat Cell Biol. 2003;5:422–426. - PubMed
-
- Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18:2670–2682. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

