Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity
- PMID: 22342841
- PMCID: PMC3299805
- DOI: 10.1016/j.immuni.2011.12.015
Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity
Abstract
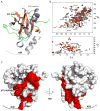
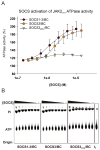
Janus kinases (JAKs) are key effectors in controlling immune responses and maintaining hematopoiesis. SOCS3 (suppressor of cytokine signaling-3) is a major regulator of JAK signaling and here we investigate the molecular basis of its mechanism of action. We found that SOCS3 bound and directly inhibited the catalytic domains of JAK1, JAK2, and TYK2 but not JAK3 via an evolutionarily conserved motif unique to JAKs. Mutation of this motif led to the formation of an active kinase that could not be inhibited by SOCS3. Surprisingly, we found that SOCS3 simultaneously bound JAK and the cytokine receptor to which it is attached, revealing how specificity is generated in SOCS action and explaining why SOCS3 inhibits only a subset of cytokines. Importantly, SOCS3 inhibited JAKs via a noncompetitive mechanism, making it a template for the development of specific and effective inhibitors to treat JAK-based immune and proliferative diseases.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Comment in
-
JAK's SOCS: a mechanism of inhibition.Immunity. 2012 Feb 24;36(2):157-9. doi: 10.1016/j.immuni.2012.01.010. Immunity. 2012. PMID: 22365659
References
-
- Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101:2271–2290. - PubMed
-
- Babon JJ, McManus EJ, Yao S, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, et al. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell. 2006;22:205–216. - PubMed
-
- Babon JJ, Yao S, DeSouza DP, Harrison CF, Fabri LJ, Liepinsh E, Scrofani SD, Baca M, Norton RS. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. Febs J. 2005;272:6120–6130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

