Down-regulation of MutS homolog 3 by hypoxia in human colorectal cancer
- PMID: 22343000
- PMCID: PMC3793328
- DOI: 10.1016/j.bbamcr.2012.01.017
Down-regulation of MutS homolog 3 by hypoxia in human colorectal cancer
Abstract
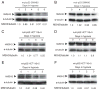
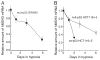
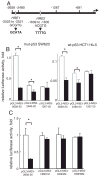
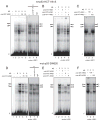
Down-regulation of hMSH3 is associated with elevated microsatellite alterations at selected tetranucleotide repeats and low levels of microsatellite instability in colorectal cancer (CRC). However, the mechanism that down-regulates hMSH3 in CRC is not known. In this study, a significant association between over-expression of glucose transporter 1, a marker for hypoxia, and down-regulation of hMSH3 in CRC tissues was observed. Therefore, we examined the effect of hypoxia on the expression of hMSH3 in human cell lines. When cells with wild type p53 (wt-p53) were exposed to hypoxia, rapid down-regulation of both hMSH2 and hMSH3 occurred. In contrast, when null or mutated p53 (null/mut-p53) cells were exposed to hypoxia, only hMSH3 was down-regulated, and at slower rate than wt-p53 cells. Using a reporter assay, we found that disruption of the two putative hypoxia response elements (HREs) located within the promoter region of the hMSH3 abrogated the suppressive effect of hypoxia on reporter activity regardless of p53 status. In an EMSA, two different forms of HIF-1α complexes that specifically bind to these HREs were detected. A larger complex containing HIF-1α predominantly bound to the HREs in hypoxic null/mut-p53 cells whereas a smaller complex predominated in wt-p53 cells. Finally, HIF-1α knockdown by siRNA significantly inhibited down-regulation of hMSH3 by hypoxia in both wt-p53 and mut-p53 cells. Taken together, our results suggest that the binding of HIF-1α complexes to HRE sites is necessary for down-regulation of hMSH3 in both wt-p53 and mut-p53 cells.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Oxidative stress induces nuclear-to-cytosol shift of hMSH3, a potential mechanism for EMAST in colorectal cancer cells.PLoS One. 2012;7(11):e50616. doi: 10.1371/journal.pone.0050616. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226332 Free PMC article.
-
HIF-1α activates hypoxia-induced BCL-9 expression in human colorectal cancer cells.Oncotarget. 2017 Apr 18;8(16):25885-25896. doi: 10.18632/oncotarget.8834. Oncotarget. 2017. PMID: 27121066 Free PMC article.
-
HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression.Mol Cell. 2005 Mar 18;17(6):793-803. doi: 10.1016/j.molcel.2005.02.015. Mol Cell. 2005. PMID: 15780936
-
Oxygen tension regulates the expression of ANK (progressive ankylosis) in an HIF-1-dependent manner in growth plate chondrocytes.J Bone Miner Res. 2009 Nov;24(11):1869-78. doi: 10.1359/jbmr.090512. J Bone Miner Res. 2009. PMID: 19419319 Free PMC article.
-
Hypoxic Modulation of HLA-G Expression through the Metabolic Sensor HIF-1 in Human Cancer Cells.J Immunol Res. 2017;2017:4587520. doi: 10.1155/2017/4587520. Epub 2017 Jul 11. J Immunol Res. 2017. PMID: 28781970 Free PMC article. Review.
Cited by
-
EMAST is a Form of Microsatellite Instability That is Initiated by Inflammation and Modulates Colorectal Cancer Progression.Genes (Basel). 2015 Mar 31;6(2):185-205. doi: 10.3390/genes6020185. Genes (Basel). 2015. PMID: 25836926 Free PMC article. Review.
-
Tumor-specific microsatellite instability: do distinct mechanisms underlie the MSI-L and EMAST phenotypes?Mutat Res. 2013 Mar-Apr;743-744:67-77. doi: 10.1016/j.mrfmmm.2012.11.003. Epub 2012 Dec 1. Mutat Res. 2013. PMID: 23206442 Free PMC article. Review.
-
Tumor hypoxia as a driving force in genetic instability.Genome Integr. 2013 Oct 24;4(1):5. doi: 10.1186/2041-9414-4-5. Genome Integr. 2013. PMID: 24152759 Free PMC article.
-
Inflammation-associated microsatellite alterations: Mechanisms and significance in the prognosis of patients with colorectal cancer.World J Gastrointest Oncol. 2018 Jan 15;10(1):1-14. doi: 10.4251/wjgo.v10.i1.1. World J Gastrointest Oncol. 2018. PMID: 29375743 Free PMC article. Review.
-
Microsatellite Alterations With Allelic Loss at 9p24.2 Signify Less-Aggressive Colorectal Cancer Metastasis.Gastroenterology. 2016 Apr;150(4):944-55. doi: 10.1053/j.gastro.2015.12.032. Epub 2016 Jan 2. Gastroenterology. 2016. PMID: 26752111 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

