Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population
- PMID: 22343293
- PMCID: PMC3318454
- DOI: 10.1128/JB.00022-12
Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population
Abstract
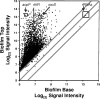
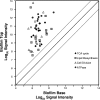
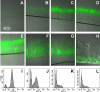
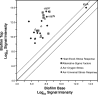
Bacteria growing in biofilms are physiologically heterogeneous, due in part to their adaptation to local environmental conditions. Here, we characterized the local transcriptome responses of Pseudomonas aeruginosa growing in biofilms by using a microarray analysis of isolated biofilm subpopulations. The results demonstrated that cells at the top of the biofilms had high mRNA abundances for genes involved in general metabolic functions, while mRNA levels for these housekeeping genes were low in cells at the bottom of the biofilms. Selective green fluorescent protein (GFP) labeling showed that cells at the top of the biofilm were actively dividing. However, the dividing cells had high mRNA levels for genes regulated by the hypoxia-induced regulator Anr. Slow-growing cells deep in the biofilms had little expression of Anr-regulated genes and may have experienced long-term anoxia. Transcripts for ribosomal proteins were associated primarily with the metabolically active cell fraction, while ribosomal RNAs were abundant throughout the biofilms, indicating that ribosomes are stably maintained even in slowly growing cells. Consistent with these results was the identification of mRNAs for ribosome hibernation factors (the rmf and PA4463 genes) at the bottom of the biofilms. The dormant biofilm cells of a P. aeruginosa Δrmf strain had decreased membrane integrity, as shown by propidium iodide staining. Using selective GFP labeling and cell sorting, we show that the dividing cells are more susceptible to killing by tobramycin and ciprofloxacin. The results demonstrate that in thick P. aeruginosa biofilms, cells are physiologically distinct spatially, with cells deep in the biofilm in a viable but antibiotic-tolerant slow-growth state.
Figures

References
-
- Alhede M, et al. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155: 3500– 3508 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

