A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens
- PMID: 22343343
- PMCID: PMC3482495
- DOI: 10.1038/nmeth.1898
A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens
Abstract
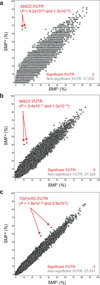
Because off-target effects hamper interpretation and validation of RNAi screen data, we developed a bioinformatics method, genome-wide enrichment of seed sequence matches (GESS), to identify candidate off-targeted transcripts in primary screening data. GESS analysis revealed a prominent off-targeted transcript in several screens, including MAD2 (MAD2L1) in a screen for genes required for the spindle assembly checkpoint. GESS analysis results can enhance the validation rate in RNAi screens.
Figures


References
ONLINE METHODS REFERENCES
-
- Benjamini Y, Hochberg Y. J Roy Statist Soc Ser B. 1995;57:289–300.
-
- Bonferroni C. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62.
-
- Holm S. Scandinavian Journal of Statistics. 1979;6:65–70.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

