CIP, a cardiac Isl1-interacting protein, represses cardiomyocyte hypertrophy
- PMID: 22343712
- PMCID: PMC3307880
- DOI: 10.1161/CIRCRESAHA.111.259663
CIP, a cardiac Isl1-interacting protein, represses cardiomyocyte hypertrophy
Abstract
Rationale: Mammalian heart has minimal regenerative capacity. In response to mechanical or pathological stress, the heart undergoes cardiac remodeling. Pressure and volume overload in the heart cause increased size (hypertrophic growth) of cardiomyocytes. Whereas the regulatory pathways that activate cardiac hypertrophy have been well-established, the molecular events that inhibit or repress cardiac hypertrophy are less known.
Objective: To identify and investigate novel regulators that modulate cardiac hypertrophy.
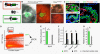
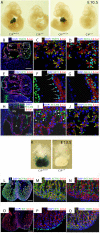
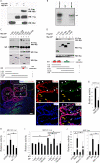
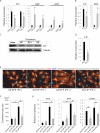
Methods and results: Here, we report the identification, characterization, and functional examination of a novel cardiac Isl1-interacting protein (CIP). CIP was identified from a bioinformatic search for novel cardiac-expressed genes in mouse embryonic hearts. CIP encodes a nuclear protein without recognizable motifs. Northern blotting, in situ hybridization, and reporter gene tracing demonstrated that CIP is highly expressed in cardiomyocytes of developing and adult hearts. Yeast two-hybrid screening identified Isl1, a LIM/homeodomain transcription factor essential for the specification of cardiac progenitor cells in the second heart field, as a cofactor of CIP. CIP directly interacted with Isl1, and we mapped the domains of these two proteins, which mediate their interaction. We show that CIP represses the transcriptional activity of Isl1 in the activation of the myocyte enhancer factor 2C. The expression of CIP was dramatically reduced in hypertrophic cardiomyocytes. Most importantly, overexpression of CIP repressed agonist-induced cardiomyocyte hypertrophy.
Conclusions: Our studies therefore identify CIP as a novel regulator of cardiac hypertrophy.
Figures


Comment in
-
Human genome organization--symbolized muscle-enriched a-type lamin-interacting protein to clear up confusion.Circ Res. 2012 Oct 12;111(9):e252; author reply e253-4. doi: 10.1161/CIRCRESAHA.112.278614. Circ Res. 2012. PMID: 23065348 No abstract available.
Similar articles
-
Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis.J Clin Invest. 2015 Nov 2;125(11):4122-34. doi: 10.1172/JCI82423. Epub 2015 Oct 5. J Clin Invest. 2015. PMID: 26436652 Free PMC article.
-
FHL2 switches MITF from activator to repressor of Erbin expression during cardiac hypertrophy.Int J Cardiol. 2015 Sep 15;195:85-94. doi: 10.1016/j.ijcard.2015.05.108. Epub 2015 May 20. Int J Cardiol. 2015. PMID: 26025865 Free PMC article.
-
Human genome organization--symbolized muscle-enriched a-type lamin-interacting protein to clear up confusion.Circ Res. 2012 Oct 12;111(9):e252; author reply e253-4. doi: 10.1161/CIRCRESAHA.112.278614. Circ Res. 2012. PMID: 23065348 No abstract available.
-
Direct nkx2-5 transcriptional repression of isl1 controls cardiomyocyte subtype identity.Stem Cells. 2015 Apr;33(4):1113-29. doi: 10.1002/stem.1923. Stem Cells. 2015. PMID: 25524439 Free PMC article.
-
The novel cardiac z-disc protein CEFIP regulates cardiomyocyte hypertrophy by modulating calcineurin signaling.J Biol Chem. 2017 Sep 15;292(37):15180-15191. doi: 10.1074/jbc.M117.786764. Epub 2017 Jul 17. J Biol Chem. 2017. PMID: 28717008 Free PMC article.
Cited by
-
MicroRNA-155 facilitates skeletal muscle regeneration by balancing pro- and anti-inflammatory macrophages.Cell Death Dis. 2016 Jun 9;7(6):e2261. doi: 10.1038/cddis.2016.165. Cell Death Dis. 2016. PMID: 27277683 Free PMC article.
-
Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis.J Clin Invest. 2015 Nov 2;125(11):4122-34. doi: 10.1172/JCI82423. Epub 2015 Oct 5. J Clin Invest. 2015. PMID: 26436652 Free PMC article.
-
Regulation of myonuclear positioning and muscle function by the skeletal muscle-specific CIP protein.Proc Natl Acad Sci U S A. 2020 Aug 11;117(32):19254-19265. doi: 10.1073/pnas.1922911117. Epub 2020 Jul 27. Proc Natl Acad Sci U S A. 2020. PMID: 32719146 Free PMC article.
-
MicroRNA-130a attenuates cardiac fibrosis after myocardial infarction through TGF-β/Smad signaling by directly targeting TGF-β receptor 1.Bioengineered. 2022 Mar;13(3):5779-5791. doi: 10.1080/21655979.2022.2033380. Bioengineered. 2022. PMID: 35188441 Free PMC article.
-
Spotlight on Isl1: A Key Player in Cardiovascular Development and Diseases.Front Cell Dev Biol. 2021 Nov 25;9:793605. doi: 10.3389/fcell.2021.793605. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34901033 Free PMC article. Review.
References
-
- Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;10(5):467–474. - PubMed
-
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9(13):1654–1666. - PubMed
-
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119(3):969. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

