Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding
- PMID: 22343733
- PMCID: PMC3419980
- DOI: 10.1038/leu.2012.49
Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding
Abstract
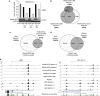
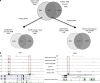
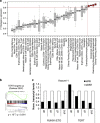
The t(8;21) translocation fuses the DNA-binding domain of the hematopoietic master regulator RUNX1 to the ETO protein. The resultant RUNX1/ETO fusion protein is a leukemia-initiating transcription factor that interferes with RUNX1 function. The result of this interference is a block in differentiation and, finally, the development of acute myeloid leukemia (AML). To obtain insights into RUNX1/ETO-dependant alterations of the epigenetic landscape, we measured genome-wide RUNX1- and RUNX1/ETO-bound regions in t(8;21) cells and assessed to what extent the effects of RUNX1/ETO on the epigenome depend on its continued expression in established leukemic cells. To this end, we determined dynamic alterations of histone acetylation, RNA Polymerase II binding and RUNX1 occupancy in the presence or absence of RUNX1/ETO using a knockdown approach. Combined global assessments of chromatin accessibility and kinetic gene expression data show that RUNX1/ETO controls the expression of important regulators of hematopoietic differentiation and self-renewal. We show that selective removal of RUNX1/ETO leads to a widespread reversal of epigenetic reprogramming and a genome-wide redistribution of RUNX1 binding, resulting in the inhibition of leukemic proliferation and self-renewal, and the induction of differentiation. This demonstrates that RUNX1/ETO represents a pivotal therapeutic target in AML.
Figures





References
-
- Bonifer C, Bowen DT. Epigenetic mechanisms regulating normal and malignant haematopoiesis: new therapeutic targets for clinical medicine. Expert Rev Mol Med. 2010;12:e6. - PubMed
-
- Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. - PubMed
-
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. - PubMed
-
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

