A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response
- PMID: 22344029
- PMCID: PMC3290715
- DOI: 10.1038/ncb2426
A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response
Abstract
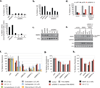
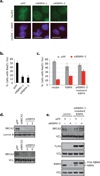
Repair of DNA double-strand breaks is critical to genomic stability and the prevention of developmental disorders and cancer. A central pathway for this repair is homologous recombination (HR). Most knowledge of HR is derived from work in prokaryotic and eukaryotic model organisms. We carried out a genome-wide siRNA-based screen in human cells. Among positive regulators of HR we identified networks of DNA-damage-response and pre-mRNA-processing proteins, and among negative regulators we identified a phosphatase network. Three candidate proteins localized to DNA lesions, including RBMX, a heterogeneous nuclear ribonucleoprotein that has a role in alternative splicing. RBMX accumulated at DNA lesions through multiple domains in a poly(ADP-ribose) polymerase 1-dependent manner and promoted HR by facilitating proper BRCA2 expression. Our screen also revealed that off-target depletion of RAD51 is a common source of RNAi false positives, raising a cautionary note for siRNA screens and RNAi-based studies of HR.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

