Identification of novel type 1 diabetes candidate genes by integrating genome-wide association data, protein-protein interactions, and human pancreatic islet gene expression
- PMID: 22344559
- PMCID: PMC3314366
- DOI: 10.2337/db11-1263
Identification of novel type 1 diabetes candidate genes by integrating genome-wide association data, protein-protein interactions, and human pancreatic islet gene expression
Abstract
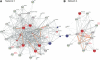
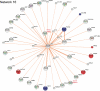
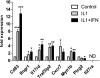
Genome-wide association studies (GWAS) have heralded a new era in susceptibility locus discovery in complex diseases. For type 1 diabetes, >40 susceptibility loci have been discovered. However, GWAS do not inevitably lead to identification of the gene or genes in a given locus associated with disease, and they do not typically inform the broader context in which the disease genes operate. Here, we integrated type 1 diabetes GWAS data with protein-protein interactions to construct biological networks of relevance for disease. A total of 17 networks were identified. To prioritize and substantiate these networks, we performed expressional profiling in human pancreatic islets exposed to proinflammatory cytokines. Three networks were significantly enriched for cytokine-regulated genes and, thus, likely to play an important role for type 1 diabetes in pancreatic islets. Eight of the regulated genes (CD83, IFNGR1, IL17RD, TRAF3IP2, IL27RA, PLCG2, MYO1B, and CXCR7) in these networks also harbored single nucleotide polymorphisms nominally associated with type 1 diabetes. Finally, the expression and cytokine regulation of these new candidate genes were confirmed in insulin-secreting INS-1 β-cells. Our results provide novel insight to the mechanisms behind type 1 diabetes pathogenesis and, thus, may provide the basis for the design of novel treatment strategies.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

