Transcriptome mapping of pAR060302, a blaCMY-2-positive broad-host-range IncA/C plasmid
- PMID: 22344651
- PMCID: PMC3346456
- DOI: 10.1128/AEM.07199-11
Transcriptome mapping of pAR060302, a blaCMY-2-positive broad-host-range IncA/C plasmid
Abstract
The multidrug resistance-encoding plasmids belonging to the IncA/C incompatibility group have recently emerged among Escherichia coli and Salmonella enterica strains in the United States. These plasmids have a unique genetic structure compared to other enterobacterial plasmid types, a broad host range, and a propensity to acquire large numbers of antimicrobial resistance genes via their accessory regions. Using E. coli strain DH5α harboring the prototype IncA/C plasmid pAR060302, we sought to define the baseline transcriptome of IncA/C plasmids under laboratory growth and in the face of selective pressure. The effects of ampicillin, florfenicol, or streptomycin exposure were compared to those on cells left untreated at logarithmic phase using Illumina platform-based RNA sequencing (RNA-Seq). Under growth in Luria-Bertani broth lacking antibiotics, much of the backbone of pAR060302 was transcriptionally inactive, including its putative transfer regions. A few plasmid backbone genes of interest were highly transcribed, including genes of a putative toxin-antitoxin system and an H-NS-like transcriptional regulator. In contrast, numerous genes within the accessory regions of pAR060302 were highly transcribed, including the resistance genes floR, bla(CMY-2), aadA, and aacA. Treatment with ampicillin or streptomycin resulted in no genes being differentially expressed compared to controls lacking antibiotics, suggesting that many of the resistance-associated genes are not differentially expressed due to exposure to these antibiotics. In contrast, florfenicol treatment resulted in the upregulation of floR and numerous chromosomal genes. Overall, the transcriptome mapping of pAR060302 suggests that it mitigates the fitness costs of carrying resistance-associated genes through global regulation with its transcriptional regulators.
Figures




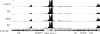
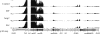
References
-
- Alonso A, Sánchez P, Martínez JL. 2001. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3:1–9 - PubMed
-
- Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74 - PubMed
-
- Blickwede M, Goethe R, Wolz C, Valentin-Weigand P, Schwarz S. 2005. Molecular basis of florfenicol-induced increase in adherence of Staphylococcus aureus strain Newman. J. Antimicrob. Chemother. 56:315–323 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

