Integrated analysis identifies a class of androgen-responsive genes regulated by short combinatorial long-range mechanism facilitated by CTCF
- PMID: 22344698
- PMCID: PMC3367180
- DOI: 10.1093/nar/gks139
Integrated analysis identifies a class of androgen-responsive genes regulated by short combinatorial long-range mechanism facilitated by CTCF
Abstract
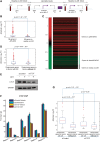
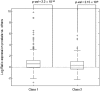
Recently, much attention has been given to elucidate how long-range gene regulation comes into play and how histone modifications and distal transcription factor binding contribute toward this mechanism. Androgen receptor (AR), a key regulator of prostate cancer, has been shown to regulate its target genes via distal enhancers, leading to the hypothesis of global long-range gene regulation. However, despite numerous flows of newly generated data, the precise mechanism with respect to AR-mediated long-range gene regulation is still largely unknown. In this study, we carried out an integrated analysis combining several types of high-throughput data, including genome-wide distribution data of H3K4 di-methylation (H3K4me2), CCCTC binding factor (CTCF), AR and FoxA1 cistrome data as well as androgen-regulated gene expression data. We found that a subset of androgen-responsive genes was significantly enriched near AR/H3K4me2 overlapping regions and FoxA1 binding sites within the same CTCF block. Importantly, genes in this class were enriched in cancer-related pathways and were downregulated in clinical metastatic versus localized prostate cancer. Our results suggest a relatively short combinatorial long-range regulation mechanism facilitated by CTCF blocking. Under such a mechanism, H3K4me2, AR and FoxA1 within the same CTCF block combinatorially regulate a subset of distally located androgen-responsive genes involved in prostate carcinogenesis.
Figures




References
-
- Hääg P, Bektic J, Bartsch G, Klocker H, Eder IE. Androgen receptor down regulation by small interference RNA induces cell growth inhibition in androgen sensitive as well as in androgen independent prostate cancer cells. J. Steroid Biochem. Mol. Biol. 2005;96:251–258. - PubMed
-
- Scher HI, Liebertz C, Kelly WK, Mazumdar M, Brett C, Schwartz L, Kolvenbag G, Shapiro L, Schwartz M. Bicalutamide for advanced prostate cancer: the natural versus treated history of disease. J. Clin. Oncol. 1997;15:2928–2938. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

