PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4
- PMID: 22345481
- PMCID: PMC3318667
- DOI: 10.1128/JVI.06973-11
PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4
Abstract
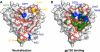
Recently, several broadly neutralizing monoclonal antibodies (bnMAbs) directed to the CD4-binding site (CD4bs) of gp120 have been isolated from HIV-1-positive donors. These include VRC01, 3BNC117, and NIH45-46, all of which are capable of neutralizing about 90% of circulating HIV-1 isolates and all of which induce conformational changes in the HIV-1 gp120 monomer similar to those induced by the CD4 receptor. In this study, we characterize PGV04 (also known as VRC-PG04), a MAb with potency and breadth that rivals those of the prototypic VRC01 and 3BNC117. When screened on a large panel of viruses, the neutralizing profile of PGV04 was distinct from those of CD4, b12, and VRC01. Furthermore, the ability of PGV04 to neutralize pseudovirus containing single alanine substitutions exhibited a pattern distinct from those of the other CD4bs MAbs. In particular, substitutions D279A, I420A, and I423A were found to abrogate PGV04 neutralization. In contrast to VRC01, PGV04 did not enhance the binding of 17b or X5 to their epitopes (the CD4-induced [CD4i] site) in the coreceptor region on the gp120 monomer. Furthermore, in contrast to CD4, none of the anti-CD4bs MAbs induced the expression of the 17b epitope on cell surface-expressed cleaved Env trimers. We conclude that potent CD4bs bnMAbs can display differences in the way they recognize and access the CD4bs and that mimicry of CD4, as assessed by inducing conformational changes in monomeric gp120 that lead to enhanced exposure of the CD4i site, is not uniquely correlated with effective neutralization at the site of CD4 binding on HIV-1.
Figures






References
-
- Burton DR, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

