The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices
- PMID: 22345483
- PMCID: PMC3318633
- DOI: 10.1128/JVI.00012-12
The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices
Abstract
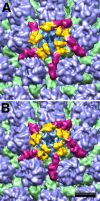
Herpesviruses have an icosahedral nucleocapsid surrounded by an amorphous tegument and a lipoprotein envelope. The tegument comprises at least 20 proteins destined for delivery into the host cell. As the tegument does not have a regular structure, the question arises of how its proteins are recruited. The herpes simplex virus 1 (HSV-1) tegument is known to contact the capsid at its vertices, and two proteins, UL36 and UL37, have been identified as candidates for this interaction. We show that the interaction is mediated exclusively by UL36. HSV-1 nucleocapsids extracted from virions shed their UL37 upon incubation at 37°C. Cryo-electron microscopy (cryo-EM) analysis of capsids with and without UL37 reveals the same penton-capping density in both cases. As no other tegument proteins are retained in significant amounts, it follows that this density feature (∼100 kDa) represents the ordered portion of UL36 (336 kDa). It binds between neighboring UL19 protrusions and to an adjacent UL17 molecule. These observations support the hypothesis that UL36 plays a major role in the tegumentation of the virion, providing a flexible scaffold to which other tegument proteins, including UL37, bind. They also indicate how sequential conformational changes in the maturing nucleocapsid control the ordered binding, first of UL25/UL17 and then of UL36.
Figures




References
-
- Baines JD. 2007. Envelopment of herpes simplex virus nucleocapsids at the inner membrane, p 144–150 In Arvin A, et al. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom
-
- Britt B. 2007. CMV maturation and egress, p 311–323 In Arvin A, et al. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

