Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis
- PMID: 22345490
- PMCID: PMC3315224
- DOI: 10.1105/tpc.111.094144
Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis
Erratum in
- Plant Cell. 2012 Mar;24(3):1301
Abstract
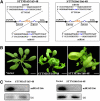
MicroRNAs (miRNAs) and other endogenous small RNAs act as sequence-specific regulators of the genome, transcriptome, and proteome in eukaryotes. The interrogation of small RNA functions requires an effective, widely applicable method to specifically block small RNA function. Here, we report the development of a highly effective technology that targets specific endogenous miRNAs or small interfering RNAs for destruction in Arabidopsis thaliana. We show that the expression of a short tandem target mimic (STTM), which is composed of two short sequences mimicking small RNA target sites, separated by a linker of an empirically determined optimal size, leads to the degradation of targeted small RNAs by small RNA degrading nucleases. The efficacy of the technology was demonstrated by the strong and specific developmental defects triggered by STTMs targeting three miRNAs and an endogenous siRNA. In summary, we developed an effective approach for the destruction of endogenous small RNAs, thereby providing a powerful tool for functional genomics of small RNA molecules in plants and potentially animals.
Figures
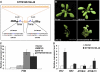
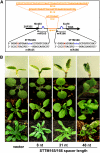

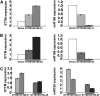
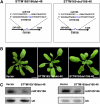



Comment in
-
A new tool for investigating small RNA function.Plant Cell. 2012 Feb;24(2):372. doi: 10.1105/tpc.112.240211. Epub 2012 Feb 21. Plant Cell. 2012. PMID: 22353372 Free PMC article. No abstract available.
References
-
- Baker C.C., Sieber P., Wellmer F., Meyerowitz E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15: 303–315 - PubMed
-
- Brosnan C.A., Voinnet O. (2009). The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 21: 416–425 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

