Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination
- PMID: 22345518
- PMCID: PMC3289885
- DOI: 10.1101/gad.176156.111
Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination
Abstract
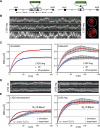
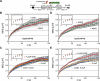
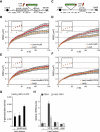
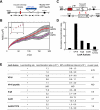
Chromatin in the interphase nucleus moves in a constrained random walk. Despite extensive study, the molecular causes of such movement and its impact on DNA-based reactions are unclear. Using high-precision live fluorescence microscopy in budding yeast, we quantified the movement of tagged chromosomal loci to which transcriptional activators or nucleosome remodeling complexes were targeted. We found that local binding of the transcriptional activator VP16, but not of the Gal4 acidic domain, enhances chromatin mobility. The increase in movement did not correlate strictly with RNA polymerase II (PolII) elongation, but could be phenocopied by targeting the INO80 remodeler to the locus. Enhanced chromatin mobility required Ino80's ATPase activity. Consistently, the INO80-dependent remodeling of nucleosomes upon transcriptional activation of the endogenous PHO5 promoter enhanced chromatin movement locally. Finally, increased mobility at a double-strand break was also shown to depend in part on the INO80 complex. This correlated with increased rates of spontaneous gene conversion. We propose that local chromatin remodeling and nucleosome eviction increase large-scale chromatin movements by enhancing the flexibility of the chromatin fiber.
Figures


References
-
- Barbaric S, Luckenbach T, Schmid A, Blaschke D, Horz W, Korber P 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J Biol Chem 282: 27610–27621 - PubMed
-
- Berg HC. 1993. Random walks in biology. Princeton University Press, Princeton, NJ.
-
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, Triezenberg SJ, Guarente L 1992. Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70: 251–265 - PubMed
-
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD 2004. Romoval of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell 14: 667–673 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
