Translocation Biosensors - Cellular System Integrators to Dissect CRM1-Dependent Nuclear Export by Chemicogenomics
- PMID: 22346706
- PMCID: PMC3274152
- DOI: 10.3390/s90705423
Translocation Biosensors - Cellular System Integrators to Dissect CRM1-Dependent Nuclear Export by Chemicogenomics
Abstract
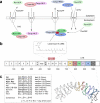
Fluorescent protein biosensors are powerful cellular systems biology tools for dissecting the complexity of cellular processes with high spatial and temporal resolution. As regulated nucleo-cytoplasmic transport is crucial for the modulation of numerous (patho)physiological cellular responses, a detailed understanding of its molecular mechanism would open up novel options for a rational manipulation of the cell. In contrast to genetic approaches, we here established and employed high-content cellular translocation biosensors applicable for dissecting nuclear export by chemicogenomics. A431 cell lines, stably expressing a translocation biosensor composed of glutathione S-transferase, GFP and a rational combination of nuclear import and export signals, were engineered by antibiotic selection and flow cytometry sorting. Using an optimized nuclear translocation algorithm, the translocation response could be robustly quantified on the Cellomics Arrayscan(®) VTI platform. Subsequent to assay optimization, the assay was developed into a higher density 384-well format high-content assay and employed for the screening of the 17K ChemBioNet compound collection. This library was selected on the basis of a genetic algorithm used to identify maximum common chemical substructures in a database of annotated bioactive molecules and hence, is well-placed in the chemical space covered by bioactive compounds. Automated multiparameter data analysis combined with visual inspection allowed us to identify and to rationally discriminate true export inhibitors from false positives, which included fluorescent compounds or cytotoxic substances that dramatically affected the cellular morphology. A total of 120 potential hit compounds were selected for Cellomics Arrayscan(®) VTI based rescreening. The export inhibitory activity of 20 compounds effective at concentrations < 25 μM were confirmed by fluorescence microscopy in several cell lines. Interestingly, kinetic analysis allowed the identification of inhibitors capable to interfere with the export receptor CRM1-mediated nuclear export not only in an irreversible, but also in a reversible fashion. In sum, exploitation of biosensor based screening allows the identification of chemicogenomic tools applicable for dissecting nucleo-cytoplasmic transport in living cells.
Keywords: Exportin 1/CRM1; HIV-1 Rev; LMB; cancer; chemical biology; import; nucleocytoplasmic transport; nucleoporin.
Figures







Similar articles
-
Translocation biosensors to study signal-specific nucleo-cytoplasmic transport, protease activity and protein-protein interactions.Traffic. 2005 Jul;6(7):594-606. doi: 10.1111/j.1600-0854.2005.00298.x. Traffic. 2005. PMID: 15941410
-
Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals.Proc Natl Acad Sci U S A. 1999 May 25;96(11):6229-34. doi: 10.1073/pnas.96.11.6229. Proc Natl Acad Sci U S A. 1999. PMID: 10339570 Free PMC article.
-
Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1.J Virol. 1998 Nov;72(11):8627-35. doi: 10.1128/JVI.72.11.8627-8635.1998. J Virol. 1998. PMID: 9765402 Free PMC article.
-
Structural Basis of Targeting the Exportin CRM1 in Cancer.Cells. 2015 Sep 21;4(3):538-68. doi: 10.3390/cells4030538. Cells. 2015. PMID: 26402707 Free PMC article. Review.
-
Nucleocytoplasmic RNA transport in retroviral replication.Results Probl Cell Differ. 2001;34:197-217. doi: 10.1007/978-3-540-40025-7_12. Results Probl Cell Differ. 2001. PMID: 11288676 Review.
Cited by
-
Disease-relevant signalling-pathways in head and neck cancer: Taspase1's proteolytic activity fine-tunes TFIIA function.Sci Rep. 2017 Nov 2;7(1):14937. doi: 10.1038/s41598-017-14814-x. Sci Rep. 2017. PMID: 29097782 Free PMC article.
-
The protein corona protects against size- and dose-dependent toxicity of amorphous silica nanoparticles.Beilstein J Nanotechnol. 2014 Aug 27;5:1380-92. doi: 10.3762/bjnano.5.151. eCollection 2014. Beilstein J Nanotechnol. 2014. PMID: 25247121 Free PMC article.
-
Trends and Advances in Electrochemiluminescence Nanobiosensors.Sensors (Basel). 2018 Jan 9;18(1):166. doi: 10.3390/s18010166. Sensors (Basel). 2018. PMID: 29315277 Free PMC article. Review.
-
Biosensors in clinical practice: focus on oncohematology.Sensors (Basel). 2013 May 14;13(5):6423-47. doi: 10.3390/s130506423. Sensors (Basel). 2013. PMID: 23673681 Free PMC article. Review.
-
Cell-based analysis of structure-function activity of threonine aspartase 1.J Biol Chem. 2011 Jan 28;286(4):3007-17. doi: 10.1074/jbc.M110.161646. Epub 2010 Nov 17. J Biol Chem. 2011. PMID: 21084304 Free PMC article.
References
-
- Giuliano K.A., Johnston P.A., Gough A., Taylor D.L. Systems cell biology based on high-content screening. Methods Enzymol. 2006;414:601–619. - PubMed
-
- Sachs K., Perez O., Pe'er D., Lauffenburger D.A., Nolan G.P. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. - PubMed
-
- Knauer S.K., Moodt S., Berg T., Liebel U., Pepperkok R., Stauber R.H. Translocation biosensors to study signal-specific nucleo-cytoplasmic transport, protease activity and protein-protein interactions. Traffic. 2005;6:594–606. - PubMed
-
- Simpson P.B., Wafford K.A. New directions in kinetic high information content assays. Drug Discov. Today. 2006;11:237–244. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

