Multi-scale modeling of HIV infection in vitro and APOBEC3G-based anti-retroviral therapy
- PMID: 22346743
- PMCID: PMC3276540
- DOI: 10.1371/journal.pcbi.1002371
Multi-scale modeling of HIV infection in vitro and APOBEC3G-based anti-retroviral therapy
Abstract
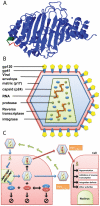
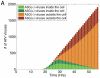
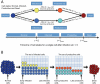
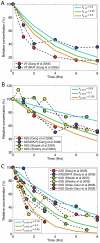
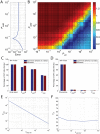
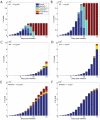
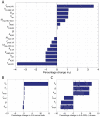
The human APOBEC3G is an innate restriction factor that, in the absence of Vif, restricts HIV-1 replication by inducing excessive deamination of cytidine residues in nascent reverse transcripts and inhibiting reverse transcription and integration. To shed light on impact of A3G-Vif interactions on HIV replication, we developed a multi-scale computational system consisting of intracellular (single-cell), cellular and extracellular (multicellular) events by using ordinary differential equations. The single-cell model describes molecular-level events within individual cells (such as production and degradation of host and viral proteins, and assembly and release of new virions), whereas the multicellular model describes the viral dynamics and multiple cycles of infection within a population of cells. We estimated the model parameters either directly from previously published experimental data or by running simulations to find the optimum values. We validated our integrated model by reproducing the results of in vitro T cell culture experiments. Crucially, both downstream effects of A3G (hypermutation and reduction of viral burst size) were necessary to replicate the experimental results in silico. We also used the model to study anti-HIV capability of several possible therapeutic strategies including: an antibody to Vif; upregulation of A3G; and mutated forms of A3G. According to our simulations, A3G with a mutated Vif binding site is predicted to be significantly more effective than other molecules at the same dose. Ultimately, we performed sensitivity analysis to identify important model parameters. The results showed that the timing of particle formation and virus release had the highest impacts on HIV replication. The model also predicted that the degradation of A3G by Vif is not a crucial step in HIV pathogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

