Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum
- PMID: 22346765
- PMCID: PMC3276549
- DOI: 10.1371/journal.pgen.1002511
Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum
Erratum in
- PLoS Genet. 2012 Mar;8(3). doi: 10.1371/annotation/b5608bc6-aa54-40a7-b246-51fa7bc4a9db doi: 10.1371/annotation/b5608bc6-aa54-40a7-b246-51fa7bc4a9db
Abstract
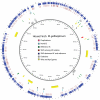
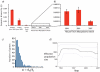
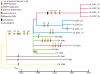
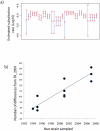
Measureable rates of genome evolution are well documented in human pathogens but are less well understood in bacterial pathogens in the wild, particularly during and after host switches. Mycoplasma gallisepticum (MG) is a pathogenic bacterium that has evolved predominantly in poultry and recently jumped to wild house finches (Carpodacus mexicanus), a common North American songbird. For the first time we characterize the genome and measure rates of genome evolution in House Finch isolates of MG, as well as in poultry outgroups. Using whole-genome sequences of 12 House Finch isolates across a 13-year serial sample and an additional four newly sequenced poultry strains, we estimate a nucleotide diversity in House Finch isolates of only ∼2% of ancestral poultry strains and a nucleotide substitution rate of 0.8-1.2×10(-5) per site per year both in poultry and in House Finches, an exceptionally fast rate rivaling some of the highest estimates reported thus far for bacteria. We also found high diversity and complete turnover of CRISPR arrays in poultry MG strains prior to the switch to the House Finch host, but after the invasion of House Finches there is progressive loss of CRISPR repeat diversity, and recruitment of novel CRISPR repeats ceases. Recent (2007) House Finch MG strains retain only ∼50% of the CRISPR repertoire founding (1994-95) strains and have lost the CRISPR-associated genes required for CRISPR function. Our results suggest that genome evolution in bacterial pathogens of wild birds can be extremely rapid and in this case is accompanied by apparent functional loss of CRISPRs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Nolan P, Hill G, Stoehr A. Sex, size, and plumage redness predict house finch survival in an epidemic. Proceedings of the Royal Society B-Biological Sciences. 1998;265:961.
-
- Dhondt AA, Dhondt KV, Hawley DM, Jennelle CS. Experimental evidence for transmission of Mycoplasma gallisepticum in house finches by fomites. Avian Pathol. 2007;36:205–208. - PubMed
-
- Dhondt AA, Tessglia DL, Slothower RL. Epidemic mycoplasmal conjunctivitis in House Finches from eastern North America. J Wildlife Dis. 1998;34:265–280. - PubMed
-
- Faustino C, Jennelle C, Connolly V, Davis A, Swarthout E, et al. Mycoplasma gallisepticum infection dynamics in a house finch population: seasonal variation in survival, encounter and transmission rate. Ecology. 2004;73:651–669.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

