Fine-Tuning Oligodendrocyte Development by microRNAs
- PMID: 22347159
- PMCID: PMC3272650
- DOI: 10.3389/fnins.2012.00013
Fine-Tuning Oligodendrocyte Development by microRNAs
Abstract
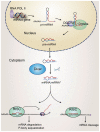
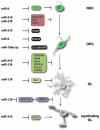
Myelination of axons by oligodendrocytes in the central nervous system is essential for normal neuronal functions. The failure of remyelination due to injury or pathological insults results in devastating demyelinating diseases. Oligodendrocytes originate in restricted regions of the embryonic ventral neural tube. After migration to populate all areas of the brain and spinal cord, oligodendrocyte precursors undergo a temporally well-defined series of molecular and structural changes, ultimately culminating in the cessation of proliferation, and the elaboration of a highly complex myelin sheath. The emergence of microRNAs (miRNAs) as potent regulators of gene expression at the posttranscriptional level has broad implications in all facets of cell biology. Recent studies have demonstrated a critical role of miRNAs in oligodendrocyte development, including cell proliferation, differentiation, and myelin formation. In this review, we will highlight and discuss the recent understanding of functional links of miRNAs to regulatory networks for central myelination, as well as perspectives on the role of miRNAs in demyelinating diseases.
Keywords: feed-back regulation; miRNAs; myelination; neural cell fate; oligodendrocyte; transcriptional control.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

