Extended co-expression of inhibitory receptors by human CD8 T-cells depending on differentiation, antigen-specificity and anatomical localization
- PMID: 22347406
- PMCID: PMC3275569
- DOI: 10.1371/journal.pone.0030852
Extended co-expression of inhibitory receptors by human CD8 T-cells depending on differentiation, antigen-specificity and anatomical localization
Abstract
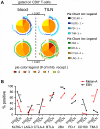
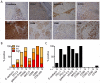
Inhibitory receptors mediate CD8 T-cell hyporesponsiveness against cancer and infectious diseases. PD-1 and CTLA-4 have been extensively studied, and blocking antibodies have already shown clinical benefit for cancer patients. Only little is known on extended co-expression of inhibitory receptors and their ligands. Here we analyzed the expression of eight inhibitory receptors by tumor-antigen specific CD8 T-cells. We found that the majority of effector T-cells simultaneously expressed four or more of the inhibitory receptors BTLA, TIM-3, LAG-3, KRLG-1, 2B4, CD160, PD-1 and CTLA-4. There were major differences depending on antigen-specificity, differentiation and anatomical localization of T-cells. On the other hand, naive T-cells were only single or double positive for BTLA and TIM-3. Extended co-expression is likely relevant for effector T-cells, as we found expression of multiple ligands in metastatic lesions of melanoma patients. Together, our data suggest that naive T-cells are primarily regulated by BTLA and TIM-3, whereas effector cells interact via larger numbers of inhibitory receptors. Blocking multiple inhibitory receptors simultaneously or sequentially may improve T-cell based therapies, but further studies are necessary to clarify the role of each receptor-ligand pair.
Conflict of interest statement
Figures


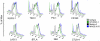

References
-
- Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175:5895–5903. - PubMed
-
- Wherry EJ, Ha SJ, Kaech SM, Haining WN, Sarkar S, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

