Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants
- PMID: 22347817
- PMCID: PMC3276548
- DOI: 10.1371/journal.pgen.1002515
Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants
Abstract
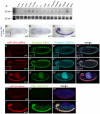
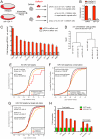
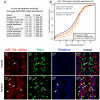
miR-124 is conserved in sequence and neuronal expression across the animal kingdom and is predicted to have hundreds of mRNA targets. Diverse defects in neural development and function were reported from miR-124 antisense studies in vertebrates, but a nematode knockout of mir-124 surprisingly lacked detectable phenotypes. To provide genetic insight from Drosophila, we deleted its single mir-124 locus and found that it is dispensable for gross aspects of neural specification and differentiation. On the other hand, we detected a variety of mutant phenotypes that were rescuable by a mir-124 genomic transgene, including short lifespan, increased dendrite variation, impaired larval locomotion, and aberrant synaptic release at the NMJ. These phenotypes reflect extensive requirements of miR-124 even under optimal culture conditions. Comparison of the transcriptomes of cells from wild-type and mir-124 mutant animals, purified on the basis of mir-124 promoter activity, revealed broad upregulation of direct miR-124 targets. However, in contrast to the proposed mutual exclusion model for miR-124 function, its functional targets were relatively highly expressed in miR-124-expressing cells and were not enriched in genes annotated with epidermal expression. A notable aspect of the direct miR-124 network was coordinate targeting of five positive components in the retrograde BMP signaling pathway, whose activation in neurons increases synaptic release at the NMJ, similar to mir-124 mutants. Derepression of the direct miR-124 target network also had many secondary effects, including over-activity of other post-transcriptional repressors and a net incomplete transition from a neuroblast to a neuronal gene expression signature. Altogether, these studies demonstrate complex consequences of miR-124 loss on neural gene expression and neurophysiology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
The miRNA pathway controls rapid changes in activity-dependent synaptic structure at the Drosophila melanogaster neuromuscular junction.PLoS One. 2013 Jul 2;8(7):e68385. doi: 10.1371/journal.pone.0068385. Print 2013. PLoS One. 2013. PMID: 23844193 Free PMC article.
-
miR-124 controls Drosophila behavior and is required for neural development.Int J Dev Neurosci. 2014 Nov;38:105-12. doi: 10.1016/j.ijdevneu.2014.08.006. Epub 2014 Aug 26. Int J Dev Neurosci. 2014. PMID: 25169673
-
Neuroligin 4 regulates synaptic growth via the bone morphogenetic protein (BMP) signaling pathway at the Drosophila neuromuscular junction.J Biol Chem. 2017 Nov 3;292(44):17991-18005. doi: 10.1074/jbc.M117.810242. Epub 2017 Sep 14. J Biol Chem. 2017. PMID: 28912273 Free PMC article.
-
The conserved miR-8/miR-200 microRNA family and their role in invertebrate and vertebrate neurogenesis.Cell Tissue Res. 2015 Jan;359(1):161-77. doi: 10.1007/s00441-014-1911-z. Epub 2014 May 30. Cell Tissue Res. 2015. PMID: 24875007 Review.
-
MiR-34 and MiR-200: Regulator of Cell Fate Plasticity and Neural Development.Neuromolecular Med. 2019 Jun;21(2):97-109. doi: 10.1007/s12017-019-08535-9. Epub 2019 Apr 8. Neuromolecular Med. 2019. PMID: 30963386 Review.
Cited by
-
The Biological Roles of microRNAs in Drosophila Development.Insects. 2024 Jun 30;15(7):491. doi: 10.3390/insects15070491. Insects. 2024. PMID: 39057224 Free PMC article. Review.
-
De novo characterization of microRNAs in oriental fruit moth Grapholita molesta and selection of reference genes for normalization of microRNA expression.PLoS One. 2017 Feb 3;12(2):e0171120. doi: 10.1371/journal.pone.0171120. eCollection 2017. PLoS One. 2017. PMID: 28158242 Free PMC article.
-
MicroRNA-Dependent Control of Sensory Neuron Function Regulates Posture Behavior in Drosophila.J Neurosci. 2021 Oct 6;41(40):8297-8308. doi: 10.1523/JNEUROSCI.0081-21.2021. Epub 2021 Aug 20. J Neurosci. 2021. PMID: 34417328 Free PMC article.
-
Approaches to manipulating microRNAs in neurogenesis.Front Neurosci. 2013 Jan 17;6:196. doi: 10.3389/fnins.2012.00196. eCollection 2012. Front Neurosci. 2013. PMID: 23335878 Free PMC article.
-
Comprehensive Assessment of the Relationship Between MicroRNA-124 and the Prognostic Significance of Cancer.Front Oncol. 2018 Jul 16;8:252. doi: 10.3389/fonc.2018.00252. eCollection 2018. Front Oncol. 2018. PMID: 30062087 Free PMC article.
References
-
- Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. - PubMed
-
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. - DOI - PMC - PubMed
-
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

