Excision of HIV-1 proviral DNA by recombinant cell permeable tre-recombinase
- PMID: 22348110
- PMCID: PMC3278460
- DOI: 10.1371/journal.pone.0031576
Excision of HIV-1 proviral DNA by recombinant cell permeable tre-recombinase
Abstract
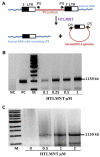
Over the previous years, comprehensive studies on antiretroviral drugs resulted in the successful introduction of highly active antiretroviral therapy (HAART) into clinical practice for treatment of HIV/AIDS. However, there is still need for new therapeutic approaches, since HAART cannot eradicate HIV-1 from the infected organism and, unfortunately, can be associated with long-term toxicity and the development of drug resistance. In contrast, novel gene therapy strategies may have the potential to reverse the infection by eradicating HIV-1. For example, expression of long terminal repeat (LTR)-specific recombinase (Tre-recombinase) has been shown to result in chromosomal excision of proviral DNA and, in consequence, in the eradication of HIV-1 from infected cell cultures. However, the delivery of Tre-recombinase currently depends on the genetic manipulation of target cells, a process that is complicating such therapeutic approaches and, thus, might be undesirable in a clinical setting. In this report we demonstrate that E.coli expressed Tre-recombinases, tagged either with the protein transduction domain (PTD) from the HIV-1 Tat trans-activator or the translocation motif (TLM) of the Hepatitis B virus PreS2 protein, were able to translocate efficiently into cells and showed significant recombination activity on HIV-1 LTR sequences. Tre activity was observed using episomal and stable integrated reporter constructs in transfected HeLa cells. Furthermore, the TLM-tagged enzyme was able to excise the full-length proviral DNA from chromosomal integration sites of HIV-1-infected HeLa and CEM-SS cells. The presented data confirm Tre-recombinase activity on integrated HIV-1 and provide the basis for the non-genetic transient application of engineered recombinases, which may be a valuable component of future HIV eradication strategies.
Conflict of interest statement
Figures
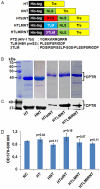
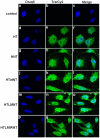
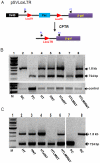

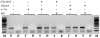
Similar articles
-
Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice.PLoS Pathog. 2013;9(9):e1003587. doi: 10.1371/journal.ppat.1003587. Epub 2013 Sep 26. PLoS Pathog. 2013. PMID: 24086129 Free PMC article.
-
Interview: HIV-1 proviral DNA excision using an evolved recombinase.J Vis Exp. 2008 Jun 16;(16):793. doi: 10.3791/793. J Vis Exp. 2008. PMID: 19066545 Free PMC article.
-
In vitro evolution and analysis of HIV-1 LTR-specific recombinases.Methods. 2011 Jan;53(1):102-9. doi: 10.1016/j.ymeth.2010.06.014. Epub 2010 Jun 26. Methods. 2011. PMID: 20600935 Review.
-
Molecular evolution of the tre recombinase.J Vis Exp. 2008 May 29;(15):791. doi: 10.3791/791. J Vis Exp. 2008. PMID: 19066582 Free PMC article.
-
Molecular biological assessment methods and understanding the course of the HIV infection.APMIS Suppl. 2003;(114):1-37. APMIS Suppl. 2003. PMID: 14626050 Review.
Cited by
-
Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice.PLoS Pathog. 2013;9(9):e1003587. doi: 10.1371/journal.ppat.1003587. Epub 2013 Sep 26. PLoS Pathog. 2013. PMID: 24086129 Free PMC article.
-
A peptide derived from laminin-γ3 reversibly impairs spermatogenesis in rats.Nat Commun. 2012;3:1185. doi: 10.1038/ncomms2171. Nat Commun. 2012. PMID: 23149730 Free PMC article.
-
DNA cleavage enzymes for treatment of persistent viral infections: recent advances and the pathway forward.Virology. 2014 Apr;454-455:353-61. doi: 10.1016/j.virol.2013.12.037. Epub 2014 Jan 31. Virology. 2014. PMID: 24485787 Free PMC article. Review.
-
Preclinical toxicity analyses of lentiviral vectors expressing the HIV-1 LTR-specific designer-recombinase Brec1.PLoS One. 2024 Mar 8;19(3):e0298542. doi: 10.1371/journal.pone.0298542. eCollection 2024. PLoS One. 2024. PMID: 38457474 Free PMC article.
-
Damaging the Integrated HIV Proviral DNA with TALENs.PLoS One. 2015 May 6;10(5):e0125652. doi: 10.1371/journal.pone.0125652. eCollection 2015. PLoS One. 2015. PMID: 25946221 Free PMC article.
References
-
- Richman DD. HIV chemotherapy. Nature. 2001;410:995–1001. - PubMed
-
- Thompson MA, Aberg JA, Cahn P, Montaner JS, Rizzardini G, et al. Antiretroviral treatment of adult HIV infection: 2010 recommendations of the International AIDS Society-USA panel. JAMA. 2010;304:321–333. - PubMed
-
- Cihlar T, Ray AS. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antiviral Res. 2010;85:39–58. - PubMed
-
- de Bethune MP. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009). Antiviral Res. 2010;85:75–90. - PubMed
-
- Martin JC, Hitchcock MJ, De Clercq E, Prusoff WH. Early nucleoside reverse transcriptase inhibitors for the treatment of HIV: a brief history of stavudine (D4T) and its comparison with other dideoxynucleosides. Antiviral Res. 2010;85:34–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

