Kinetics of DNA methylation inheritance by the Dnmt1-including complexes during the cell cycle
- PMID: 22348533
- PMCID: PMC3307489
- DOI: 10.1186/1747-1028-7-5
Kinetics of DNA methylation inheritance by the Dnmt1-including complexes during the cell cycle
Abstract
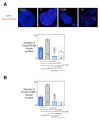
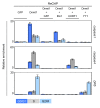
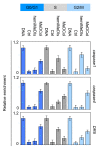
Background: The clonal transmission of lineage-specific DNA methylation patterns in a mammalian genome during the cellular division is a crucial biological process controlled by the DNA methyltransferase Dnmt1, mainly. To investigate possible dynamic mechanisms of DNA methylation inheritance during the cell cycle, we used a Proximity Ligation In Situ Assay (P-LISA) to analyze the kinetic of formation and DNA recruitment of Dnmt1-including complexes.
Results: P-LISA, sequential chromatin immunoprecipitation and quantitative methylation specific PCR revealed that the Dnmt1/PCNA/UHRF1-including complexes are mainly formed and recruited on DNA during the S-phase of cell cycle, while the formation and the DNA recruitment of several Dnmt1/transcription factors-including complexes are not S-phase dependent but are G0/G1 and/or G2/M phases dependent.
Conclusion: Our data confirm that DNA methylation inheritance occurs in S-phase, and demonstrate that DNA methylation inheritance can also occur in G0/G1 and G2/M phases of the cell cycle.
Figures


Similar articles
-
Dnmt1/Transcription factor interactions: an alternative mechanism of DNA methylation inheritance.Genes Cancer. 2010 May;1(5):434-43. doi: 10.1177/1947601910373794. Genes Cancer. 2010. PMID: 21779454 Free PMC article.
-
SET8 prevents excessive DNA methylation by methylation-mediated degradation of UHRF1 and DNMT1.Nucleic Acids Res. 2019 Sep 26;47(17):9053-9068. doi: 10.1093/nar/gkz626. Nucleic Acids Res. 2019. PMID: 31400111 Free PMC article.
-
DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation.Hum Mol Genet. 2017 Apr 15;26(8):1522-1534. doi: 10.1093/hmg/ddx057. Hum Mol Genet. 2017. PMID: 28334952 Free PMC article.
-
Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review.
-
Recruitment of Dnmt1 roles of the SRA protein Np95 (Uhrf1) and other factors.Prog Mol Biol Transl Sci. 2011;101:289-310. doi: 10.1016/B978-0-12-387685-0.00008-1. Prog Mol Biol Transl Sci. 2011. PMID: 21507355 Review.
Cited by
-
Passive DNA demethylation preferentially up-regulates pluripotency-related genes and facilitates the generation of induced pluripotent stem cells.J Biol Chem. 2017 Nov 10;292(45):18542-18555. doi: 10.1074/jbc.M117.810457. Epub 2017 Sep 18. J Biol Chem. 2017. PMID: 28924038 Free PMC article.
-
Mechanisms of establishment and functional significance of DNA demethylation during erythroid differentiation.Blood Adv. 2018 Aug 14;2(15):1833-1852. doi: 10.1182/bloodadvances.2018015651. Blood Adv. 2018. PMID: 30061308 Free PMC article.
-
Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma.Clin Epigenetics. 2018 Feb 9;10:17. doi: 10.1186/s13148-018-0450-y. eCollection 2018. Clin Epigenetics. 2018. PMID: 29449903 Free PMC article. Review.
-
ncRNAs-mediated overexpression of TET3 predicts unfavorable prognosis and correlates with immunotherapy efficacy in breast cancer.Heliyon. 2024 Jan 18;10(3):e24855. doi: 10.1016/j.heliyon.2024.e24855. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38318018 Free PMC article.
-
DNA methylation is stable during replication and cell cycle arrest.Sci Rep. 2015 Dec 9;5:17911. doi: 10.1038/srep17911. Sci Rep. 2015. PMID: 26648411 Free PMC article.
References
-
- Hervouet E, Debien E, Cheray M, Hulin P, Loussouarn D, Martin SA, Vallette FM, Cartron PF. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis by inducing genome and gene-specific hypomethylations and chromosomal instability. PLoS One. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

