GFP tagging sheds light on protein translocation: implications for key methods in cell biology
- PMID: 22349212
- PMCID: PMC11115126
- DOI: 10.1007/s00018-012-0932-6
GFP tagging sheds light on protein translocation: implications for key methods in cell biology
Abstract
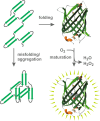
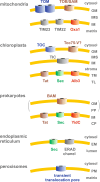
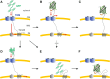
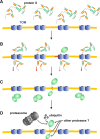
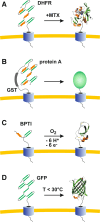
Green fluorescent protein (GFP) is a powerful tool for studying gene expression, protein localization, protein-protein interactions, calcium concentrations, and redox potentials owing to its intrinsic fluorescence. However, GFP not only contains a chromophore but is also tightly folded in a temperature-dependent manner. The latter property of GFP has recently been exploited (1) to characterize the translocase of the outer mitochondrial membrane and (2) to discriminate between protein transport across and into biomembranes in vivo. I therefore suggest that GFP could be a valuable tool for the general analysis of protein transport machineries and pathways in a variety of organisms. Moreover, results from such studies could be important for the interpretation and optimization of classical experiments using GFP tagging.
Figures
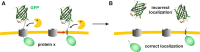
Similar articles
-
Facile manipulation of protein localization in fission yeast through binding of GFP-binding protein to GFP.J Cell Sci. 2017 Mar 1;130(5):1003-1015. doi: 10.1242/jcs.198457. Epub 2017 Jan 12. J Cell Sci. 2017. PMID: 28082423
-
Green fluorescent protein-tagging reduces the nucleocytoplasmic shuttling specifically of unphosphorylated STAT1.FEBS J. 2007 Feb;274(3):815-26. doi: 10.1111/j.1742-4658.2006.05626.x. FEBS J. 2007. PMID: 17288561
-
Analysis of DNA content and green fluorescent protein expression.Curr Protoc Cytom. 2001 May;Chapter 7:Unit 7.16. doi: 10.1002/0471142956.cy0716s16. Curr Protoc Cytom. 2001. PMID: 18770728
-
Green fluorescent protein based pH indicators for in vivo use: a review.Anal Bioanal Chem. 2009 Feb;393(4):1107-22. doi: 10.1007/s00216-008-2515-9. Epub 2008 Nov 26. Anal Bioanal Chem. 2009. PMID: 19034433 Review.
-
Fluorescent proteins for live cell imaging: opportunities, limitations, and challenges.IUBMB Life. 2009 Nov;61(11):1029-42. doi: 10.1002/iub.256. IUBMB Life. 2009. PMID: 19859977 Review.
Cited by
-
No need for labels: the autofluorescence of Leishmania tarentolae mitochondria and the necessity of negative controls.PLoS One. 2012;7(10):e47641. doi: 10.1371/journal.pone.0047641. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077654 Free PMC article.
-
Tamoxifen induces protection against manganese toxicity by REST upregulation via the ER-α/Wnt/β-catenin pathway in neuronal cells.J Biol Chem. 2025 Jun;301(6):108529. doi: 10.1016/j.jbc.2025.108529. Epub 2025 Apr 23. J Biol Chem. 2025. PMID: 40280417 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

