Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase
- PMID: 22349225
- PMCID: PMC3282621
- DOI: 10.1107/S090744491200073X
Structural basis of the substrate specificity of Bacillus cereus adenosine phosphorylase
Abstract
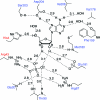
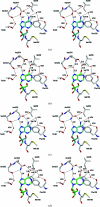
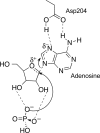
Purine nucleoside phosphorylases catalyze the phosphorolytic cleavage of the glycosidic bond of purine (2'-deoxy)nucleosides, generating the corresponding free base and (2'-deoxy)-ribose 1-phosphate. Two classes of PNPs have been identified: homotrimers specific for 6-oxopurines and homohexamers that accept both 6-oxopurines and 6-aminopurines. Bacillus cereus adenosine phosphorylase (AdoP) is a hexameric PNP; however, it is highly specific for 6-aminopurines. To investigate the structural basis for the unique substrate specificity of AdoP, the active-site mutant D204N was prepared and kinetically characterized and the structures of the wild-type protein and the D204N mutant complexed with adenosine and sulfate or with inosine and sulfate were determined at high resolution (1.2-1.4 Å). AdoP interacts directly with the preferred substrate through a hydrogen-bond donation from the catalytically important residue Asp204 to N7 of the purine base. Comparison with Escherichia coli PNP revealed a more optimal orientation of Asp204 towards N7 of adenosine and a more closed active site. When inosine is bound, two water molecules are interposed between Asp204 and the N7 and O6 atoms of the nucleoside, thus allowing the enzyme to find alternative but less efficient ways to stabilize the transition state. The mutation of Asp204 to asparagine led to a significant decrease in catalytic efficiency for adenosine without affecting the efficiency of inosine cleavage.
Figures


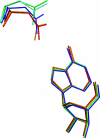
Similar articles
-
Computer Simulations Reveal Substrate Specificity of Glycosidic Bond Cleavage in Native and Mutant Human Purine Nucleoside Phosphorylase.Biochemistry. 2016 Apr 12;55(14):2153-62. doi: 10.1021/acs.biochem.5b01347. Epub 2016 Mar 29. Biochemistry. 2016. PMID: 26985580
-
Purine nucleoside phosphorylase. 1. Structure-function studies.Biochemistry. 1997 Sep 30;36(39):11725-34. doi: 10.1021/bi961969w. Biochemistry. 1997. PMID: 9305962
-
Open and closed conformation of the E. coli purine nucleoside phosphorylase active center and implications for the catalytic mechanism.J Mol Biol. 2002 Jan 18;315(3):351-71. doi: 10.1006/jmbi.2001.5211. J Mol Biol. 2002. PMID: 11786017
-
The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology.Structure. 1997 Oct 15;5(10):1373-83. doi: 10.1016/s0969-2126(97)00287-6. Structure. 1997. PMID: 9351810
-
Structural analyses reveal two distinct families of nucleoside phosphorylases.Biochem J. 2002 Jan 1;361(Pt 1):1-25. doi: 10.1042/0264-6021:3610001. Biochem J. 2002. PMID: 11743878 Free PMC article. Review.
Cited by
-
Unique substrate specificity of purine nucleoside phosphorylases from Thermus thermophilus.Extremophiles. 2013 May;17(3):505-14. doi: 10.1007/s00792-013-0535-7. Epub 2013 Apr 2. Extremophiles. 2013. PMID: 23546840
-
The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.PLoS One. 2018 Sep 7;13(9):e0203532. doi: 10.1371/journal.pone.0203532. eCollection 2018. PLoS One. 2018. PMID: 30192840 Free PMC article.
-
Crystal structure of Escherichia coli purine nucleoside phosphorylase in complex with 7-deazahypoxanthine.Acta Crystallogr F Struct Biol Commun. 2018 Jun 1;74(Pt 6):355-362. doi: 10.1107/S2053230X18006337. Epub 2018 May 23. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 29870020 Free PMC article.
-
Functional and Structural Characterization of Purine Nucleoside Phosphorylase from Kluyveromyces lactis and Its Potential Applications in Reducing Purine Content in Food.PLoS One. 2016 Oct 21;11(10):e0164279. doi: 10.1371/journal.pone.0164279. eCollection 2016. PLoS One. 2016. PMID: 27768715 Free PMC article.
-
Pediococcus acidilactici GR-5 alleviates hyperuricemia by degrading purine nucleosides and improving gut microbiota metabolism.NPJ Sci Food. 2025 Aug 23;9(1):183. doi: 10.1038/s41538-025-00556-y. NPJ Sci Food. 2025. PMID: 40849527 Free PMC article.
References
-
- Adams, P. D. et al. (2011). Methods, 55, 94–106. - PubMed
-
- Appleby, T. C., Mathews, I. I., Porcelli, M., Cacciapuoti, G. & Ealick, S. E. (2001). J. Biol. Chem. 276, 39232–39242. - PubMed
-
- Atlas, R. M. (1998). Crit. Rev. Microbiol. 24, 157–168. - PubMed
-
- Bennett, E. M., Li, C., Allan, P. W., Parker, W. B. & Ealick, S. E. (2003). J. Biol. Chem. 278, 47110–47118. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

