Characterization, high-resolution mapping and differential expression of three homologous PAL genes in Coffea canephora Pierre (Rubiaceae)
- PMID: 22349733
- PMCID: PMC3382651
- DOI: 10.1007/s00425-012-1613-2
Characterization, high-resolution mapping and differential expression of three homologous PAL genes in Coffea canephora Pierre (Rubiaceae)
Abstract
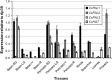
Phenylalanine ammonia lyase (PAL) is the first entry enzyme of the phenylpropanoid pathway producing phenolics, widespread constituents of plant foods and beverages, including chlorogenic acids, polyphenols found at remarkably high levels in the coffee bean and long recognized as powerful antioxidants. To date, whereas PAL is generally encoded by a small gene family, only one gene has been characterized in Coffea canephora (CcPAL1), an economically important species of cultivated coffee. In this study, a molecular- and bioinformatic-based search for CcPAL1 paralogues resulted successfully in identifying two additional genes, CcPAL2 and CcPAL3, presenting similar genomic structures and encoding proteins with close sequences. Genetic mapping helped position each gene in three different coffee linkage groups, CcPAL2 in particular, located in a coffee genome linkage group (F) which is syntenic to a region of Tomato Chromosome 9 containing a PAL gene. These results, combined with a phylogenetic study, strongly suggest that CcPAL2 may be the ancestral gene of C. canephora. A quantitative gene expression analysis was also conducted in coffee tissues, showing that all genes are transcriptionally active, but they present distinct expression levels and patterns. We discovered that CcPAL2 transcripts appeared predominantly in flower, fruit pericarp and vegetative/lignifying tissues like roots and branches, whereas CcPAL1 and CcPAL3 were highly expressed in immature fruit. This is the first comprehensive study dedicated to PAL gene family characterization in coffee, allowing us to advance functional studies which are indispensable to learning to decipher what role this family plays in channeling the metabolism of coffee phenylpropanoids.
Figures



References
-
- Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou JP, Ullmann P, Pauly M, Lapierre C, Werk-Reichhart D. A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 2006;140:30–48. doi: 10.1104/pp.105.069690. - DOI - PMC - PubMed
-
- Bazzano LA, He J, Ogden LG, Loria CM, Vupputuri S, Myers L, Whelton PK. Fruit and vegetable intake and risk of cardiovascular disease in US adults: the first National Health and Nutrition Examination Survey Epidemiologic Follow-up Study. Am J Clin Nutr. 2002;76:93–99. - PubMed
-
- Bertrand C, Noirot M, Doulbeau S, de Kochko A, Hamon S, Campa C. Chlorogenic acid content swap during fruit maturation in Coffea pseudozanguebariae. Qualitative comparison with leaves. Plant Sci. 2003;165:1355–1361. doi: 10.1016/j.plantsci.2003.07.002. - DOI
-
- Bomal C, Bedon F, Caron S, Mansfiled SD, Levasseur C, Cooke JE, Blais S, Tremblay L, Morency MJ, Pavy N, Grima-Pettenati J, Séguin A, Mackay J. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot. 2008;59:3925–3939. doi: 10.1093/jxb/ern234. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

