Cellular responses to misfolded proteins and protein aggregates
- PMID: 22350905
- PMCID: PMC4445682
- DOI: 10.1007/978-1-61779-474-2_32
Cellular responses to misfolded proteins and protein aggregates
Abstract
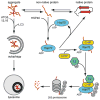
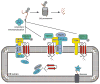
Maintenance of the proteome is a major homeostatic task of the cell and disregulation of protein homeostasis can be deadly. The accumulation of different forms of misfolded protein can perturb protein homeostasis and cause extensive cell and tissue damage. The cell has various quality control systems to help prevent the accumulation of misfolded proteins and the complexity of the different mechanisms that have evolved is bewildering. The first order of business for all quality control systems is recognition of misfolded proteins, which is followed by a triage decision. In many cases, modular molecular chaperones function in different assemblies with degradatory or folding co-factors to direct a misfolded protein toward continued life or death. Herein, an overview of quality control mechanisms that triage soluble cytosolic proteins, protein aggregates, and ER-associated proteins is presented.
Figures
References
-
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. - PubMed
-
- Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. - PubMed
-
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. - PubMed
-
- Schubert U, Anton LC, Gibbs J, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

