High dimensional and high resolution pulse sequences for backbone resonance assignment of intrinsically disordered proteins
- PMID: 22350953
- PMCID: PMC3315646
- DOI: 10.1007/s10858-012-9613-x
High dimensional and high resolution pulse sequences for backbone resonance assignment of intrinsically disordered proteins
Abstract
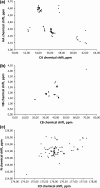
Four novel 5D (HACA(N)CONH, HNCOCACB, (HACA)CON(CA)CONH, (H)NCO(NCA)CONH), and one 6D ((H)NCO(N)CACONH) NMR pulse sequences are proposed. The new experiments employ non-uniform sampling that enables achieving high resolution in indirectly detected dimensions. The experiments facilitate resonance assignment of intrinsically disordered proteins. The novel pulse sequences were successfully tested using δ subunit (20 kDa) of Bacillus subtilis RNA polymerase that has an 81-amino acid disordered part containing various repetitive sequences.
Figures







References
-
- Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R. 13C detected protonless NMR spectroscopy of proteins in solution. Prog Nucl Magn Reson Spectrosc. 2006;48:25–45. doi: 10.1016/j.pnmrs.2005.09.002. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

