Transcriptome characterization and sequencing-based identification of salt-responsive genes in Millettia pinnata, a semi-mangrove plant
- PMID: 22351699
- PMCID: PMC3325082
- DOI: 10.1093/dnares/dss004
Transcriptome characterization and sequencing-based identification of salt-responsive genes in Millettia pinnata, a semi-mangrove plant
Abstract
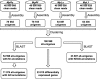
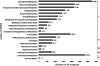
Semi-mangroves form a group of transitional species between glycophytes and halophytes, and hold unique potential for learning molecular mechanisms underlying plant salt tolerance. Millettia pinnata is a semi-mangrove plant that can survive a wide range of saline conditions in the absence of specialized morphological and physiological traits. By employing the Illumina sequencing platform, we generated ~192 million short reads from four cDNA libraries of M. pinnata and processed them into 108,598 unisequences with a high depth of coverage. The mean length and total length of these unisequences were 606 bp and 65.8 Mb, respectively. A total of 54,596 (50.3%) unisequences were assigned Nr annotations. Functional classification revealed the involvement of unisequences in various biological processes related to metabolism and environmental adaptation. We identified 23,815 candidate salt-responsive genes with significantly differential expression under seawater and freshwater treatments. Based on the reverse transcription-polymerase chain reaction (RT-PCR) and real-time PCR analyses, we verified the changes in expression levels for a number of candidate genes. The functional enrichment analyses for the candidate genes showed tissue-specific patterns of transcriptome remodelling upon salt stress in the roots and the leaves. The transcriptome of M. pinnata will provide valuable gene resources for future application in crop improvement. In addition, this study sets a good example for large-scale identification of salt-responsive genes in non-model organisms using the sequencing-based approach.
Figures
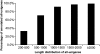
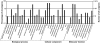

References
-
- Flowers T.J. Improving crop salt tolerance. J. Exp. Bot. 2004;55:307–19. doi:10.1093/jxb/erh003. - DOI - PubMed
-
- Hasegawa P.M., Bressan R.A., Zhu J.K., Bohnert H. J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:463–99. doi:10.1146/annurev.arplant.51.1.463. - DOI - PubMed
-
- Zhu J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–73. doi:10.1146/annurev.arplant.53.091401.143329. - DOI - PMC - PubMed
-
- Seki M., Narusaka M., Ishida J., et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–92. doi:10.1046/j.1365-313X.2002.01359.x. - DOI - PubMed
-
- Jiang Y.Q., Deyholos M.K. Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol. 2006;6:25. doi:10.1186/1471-2229-6-25. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

