Improved acid stress survival of Lactococcus lactis expressing the histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524
- PMID: 22351775
- PMCID: PMC3322857
- DOI: 10.1074/jbc.M111.330704
Improved acid stress survival of Lactococcus lactis expressing the histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524
Abstract
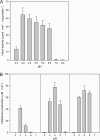
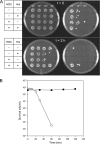
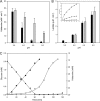
Degradative amino acid decarboxylation pathways in bacteria generate secondary metabolic energy and provide resistance against acid stress. The histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524 was functionally expressed in the heterologous host Lactococcus lactis NZ9000, and the benefits of the newly acquired pathway for the host were analyzed. During growth in M17 medium in the pH range of 5-6.5, a small positive effect was observed on the biomass yield in batch culture, whereas no growth rate enhancement was evident. In contrast, a strong benefit for the engineered L. lactis strain was observed in acid stress survival. In the presence of histidine, the pathway enabled cells to survive at pH values as low as 3 for at least 2 h, conditions under which the host cells were rapidly dying. The flux through the histidine decarboxylation pathway in cells grown at physiological pH was under strict control of the electrochemical proton gradient (pmf) across the membrane. Ionophores that dissipated the membrane potential (ΔΨ) and/or the pH gradient (ΔpH) strongly increased the flux, whereas the presence of glucose almost completely inhibited the flux. Control of the pmf over the flux was exerted by both ΔΨ and ΔpH and was distributed over the transporter HdcP and the decarboxylase HdcA. The control allowed for a synergistic effect between the histidine decarboxylation and glycolytic pathways in acid stress survival. In a narrow pH range around 2.5 the synergism resulted in a 10-fold higher survival rate.
Figures





References
-
- Abe K., Hayashi H., Maloney P. C. (1996) Exchange of aspartate and alanine. Mechanism for development of a proton-motive force in bacteria. J. Biol. Chem. 271, 3079–3084 - PubMed
-
- Pereira C. I., San Romão M. V., Lolkema J. S., Crespo M. T. (2009) Weissella halotolerans W22 combines arginine deiminase and ornithine decarboxylation pathways and converts arginine to putrescine. J. Appl. Microbiol. 107, 1894–1902 - PubMed
-
- Soksawatmaekhin W., Kuraishi A., Sakata K., Kashiwagi K., Igarashi K. (2004) Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 51, 1401–1412 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

