The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo
- PMID: 22353577
- PMCID: PMC3320176
- DOI: 10.1104/pp.112.193300
The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo
Abstract
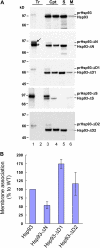
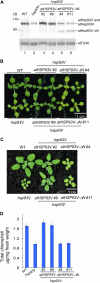
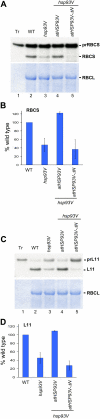
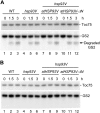
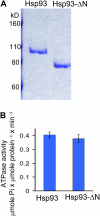
Chloroplast 93-kD heat shock protein (Hsp93/ClpC), an Hsp100 family member, is suggested to have various functions in chloroplasts, including serving as the regulatory chaperone for the ClpP protease in the stroma and acting as a motor component of the protein translocon at the envelope. Indeed, although Hsp93 is a soluble stromal protein, a portion of it is associated with the inner envelope membrane. The mechanism and functional significance of this Hsp93 membrane association have not been determined. Here, we mapped the region important for Hsp93 membrane association by creating various deletion constructs and found that only the construct with the amino-terminal domain deleted, Hsp93-ΔN, had reduced membrane association. When transformed into Arabidopsis (Arabidopsis thaliana), most atHsp93V-ΔN proteins did not associate with membranes and atHsp93V-ΔΝ failed to complement the pale-green and protein import-defective phenotypes of an hsp93V knockout mutant. The residual atHsp93V-ΔN at the membranes had further reduced association with the central protein translocon component Tic110. However, the degradation of chloroplast glutamine synthetase, a potential substrate for the ClpP protease, was not affected in the hsp93V mutant or in the atHSP93V-ΔN transgenic plants. Hsp93-ΔN also had the same ATPase activity as that of full-length Hsp93. These data suggest that the association of Hsp93 with the inner envelope membrane through its amino-terminal domain is important for the functions of Hsp93 in vivo.
Figures



References
-
- Chung MH, Chen MK, Pan SM. (2000) Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res 9: 471–476 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

