A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1-mediated transcriptional activation
- PMID: 22353606
- PMCID: PMC3297495
- DOI: 10.1186/1746-4811-8-6
A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1-mediated transcriptional activation
Abstract
Background: Arabidopsis NPR1 is a master regulator of systemic acquired resistance. NPR1 binds to TGA transcription factors and functions as a transcriptional co-activator. In rice, NH1/OsNPR1 functions to enhance innate immunity. NRR disrupts NH1 function, when over-expressed.
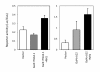
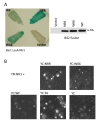
Results: We have established a rice transient protoplast assay to demonstrate that NH1 is a transcriptional co-activator and that NRR represses NH1-mediated activation. We identified three NRR homologues (RH1, RH2, and RH3). RH1 and RH3, but not RH2, also effectively repress NH1-mediated transcriptional activation. NRR, RH1, RH2, and RH3 share sequence similarity in a region beyond the previously identified NPR1-interacting domain. This region is required for strong interaction with NH1. A double point mutation, W66A/F70A, in this novel NH1-interacting domain severely reduces interaction with NH1. Mutation W66A/F70A also greatly reduces the ability of NRR to repress NH1-mediated activation. RH2 carries a deviation (amino acids AV) in this region as compared to consensus sequences (amino acids ED) among NRR, RH1, and RH3. A substitution (AV to ED) in RH2 results in strong binding of mutant RH2ED to NH1 and effective repression of NH1-mediated activation.
Conclusions: The protoplast-based transient system can be used to dissect protein domains associated with their functions. Our results demonstrate that the ability of NRR and its homologues to repress NH1-mediated transcriptional activation is tightly correlated with their ability to bind to NH1. Furthermore, a sequence is identified as a novel NH1-interacting domain. Importantly, this novel sequence is widely present in plant species, from cereals to castor bean plants, to poplar trees, to Arabidopsis, indicating its significance in plants.
Figures



Similar articles
-
A Genetic Screen Identifies a Requirement for Cysteine-Rich-Receptor-Like Kinases in Rice NH1 (OsNPR1)-Mediated Immunity.PLoS Genet. 2016 May 13;12(5):e1006049. doi: 10.1371/journal.pgen.1006049. eCollection 2016 May. PLoS Genet. 2016. PMID: 27176732 Free PMC article.
-
Interaction specificity and coexpression of rice NPR1 homologs 1 and 3 (NH1 and NH3), TGA transcription factors and Negative Regulator of Resistance (NRR) proteins.BMC Genomics. 2014 Jun 11;15(1):461. doi: 10.1186/1471-2164-15-461. BMC Genomics. 2014. PMID: 24919709 Free PMC article.
-
Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1.Plant J. 2005 Sep;43(5):623-35. doi: 10.1111/j.1365-313X.2005.02485.x. Plant J. 2005. PMID: 16115061
-
Strong suppression of systemic acquired resistance in Arabidopsis by NRR is dependent on its ability to interact with NPR1 and its putative repression domain.Mol Plant. 2008 May;1(3):552-9. doi: 10.1093/mp/ssn017. Epub 2008 Apr 22. Mol Plant. 2008. PMID: 19825560
-
Development of disease-resistant rice using regulatory components of induced disease resistance.Front Plant Sci. 2014 Nov 13;5:630. doi: 10.3389/fpls.2014.00630. eCollection 2014. Front Plant Sci. 2014. PMID: 25431577 Free PMC article. Review.
Cited by
-
The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality.Nat Genet. 2015 Aug;47(8):949-54. doi: 10.1038/ng.3352. Epub 2015 Jul 6. Nat Genet. 2015. PMID: 26147620
-
Monitoring of Rice Transcriptional Responses to Contrasted Colonizing Patterns of Phytobeneficial Burkholderia s.l. Reveals a Temporal Shift in JA Systemic Response.Front Plant Sci. 2019 Sep 24;10:1141. doi: 10.3389/fpls.2019.01141. eCollection 2019. Front Plant Sci. 2019. PMID: 31608089 Free PMC article.
-
A Genetic Screen Identifies a Requirement for Cysteine-Rich-Receptor-Like Kinases in Rice NH1 (OsNPR1)-Mediated Immunity.PLoS Genet. 2016 May 13;12(5):e1006049. doi: 10.1371/journal.pgen.1006049. eCollection 2016 May. PLoS Genet. 2016. PMID: 27176732 Free PMC article.
-
The Arabidopsis NIMIN proteins affect NPR1 differentially.Front Plant Sci. 2013 Apr 12;4:88. doi: 10.3389/fpls.2013.00088. eCollection 2013. Front Plant Sci. 2013. PMID: 23630533 Free PMC article.
-
High-quality Gossypium hirsutum and Gossypium barbadense genome assemblies reveal the landscape and evolution of centromeres.Plant Commun. 2024 Feb 12;5(2):100722. doi: 10.1016/j.xplc.2023.100722. Epub 2023 Sep 22. Plant Commun. 2024. PMID: 37742072 Free PMC article.
References
-
- Friedrich L, Lawton K, Ruess W, Masner P, Speckner N, Gt Rella M, Meier B, Dinher S, Staub T, Uknes S, Metraux J-P, Kessman H, Ryals JA. benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996;9:61–70.
-
- Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

