Stochastic signalling rewires the interaction map of a multiple feedback network during yeast evolution
- PMID: 22353713
- PMCID: PMC3293423
- DOI: 10.1038/ncomms1687
Stochastic signalling rewires the interaction map of a multiple feedback network during yeast evolution
Abstract
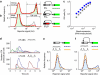
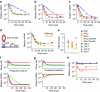
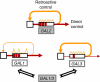
During evolution, genetic networks are rewired through strengthening or weakening their interactions to develop new regulatory schemes. In the galactose network, the GAL1/GAL3 paralogues and the GAL2 gene enhance their own expression mediated by the Gal4p transcriptional activator. The wiring strength in these feedback loops is set by the number of Gal4p binding sites. Here we show using synthetic circuits that multiplying the binding sites increases the expression of a gene under the direct control of an activator, but this enhancement is not fed back in the circuit. The feedback loops are rather activated by genes that have frequent stochastic bursts and fast RNA decay rates. In this way, rapid adaptation to galactose can be triggered even by weakly expressed genes. Our results indicate that nonlinear stochastic transcriptional responses enable feedback loops to function autonomously, or contrary to what is dictated by the strength of interactions enclosing the circuit.
Figures




Similar articles
-
Stochastic galactokinase expression underlies GAL gene induction in a GAL3 mutant of Saccharomyces cerevisiae.FEBS J. 2014 Apr;281(7):1798-817. doi: 10.1111/febs.12741. FEBS J. 2014. PMID: 24785355
-
The regulatory roles of the galactose permease and kinase in the induction response of the GAL network in Saccharomyces cerevisiae.J Biol Chem. 2006 May 12;281(19):13485-13492. doi: 10.1074/jbc.M512317200. Epub 2006 Mar 7. J Biol Chem. 2006. PMID: 16524886
-
Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p.Science. 1996 Jun 14;272(5268):1662-5. doi: 10.1126/science.272.5268.1662. Science. 1996. PMID: 8658143
-
Epigenetics of the yeast galactose genetic switch.J Biosci. 2009 Oct;34(4):513-22. doi: 10.1007/s12038-009-0070-y. J Biosci. 2009. PMID: 19920337 Review.
-
Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction.Mol Microbiol. 2001 Jun;40(5):1059-66. doi: 10.1046/j.1365-2958.2001.02421.x. Mol Microbiol. 2001. PMID: 11401712 Review.
Cited by
-
Multi-component gene network design as a survival strategy in diverse environments.BMC Syst Biol. 2018 Sep 26;12(1):85. doi: 10.1186/s12918-018-0609-3. BMC Syst Biol. 2018. PMID: 30257679 Free PMC article.
-
Comparison between Effects of Retroactivity and Resource Competition upon Change in Downstream Reporter Genes of Synthetic Genetic Circuits.Life (Basel). 2019 Mar 26;9(1):30. doi: 10.3390/life9010030. Life (Basel). 2019. PMID: 30917535 Free PMC article.
-
A unified design space of synthetic stripe-forming networks.Nat Commun. 2014 Sep 23;5:4905. doi: 10.1038/ncomms5905. Nat Commun. 2014. PMID: 25247316 Free PMC article.
-
Cross-feeding promotes heterogeneity within yeast cell populations.Nat Commun. 2024 Jan 10;15(1):418. doi: 10.1038/s41467-023-44623-y. Nat Commun. 2024. PMID: 38200012 Free PMC article.
-
A semi-synthetic regulon enables rapid growth of yeast on xylose.Nat Commun. 2018 Mar 26;9(1):1233. doi: 10.1038/s41467-018-03645-7. Nat Commun. 2018. PMID: 29581426 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

