Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus
- PMID: 22354958
- PMCID: PMC3374389
- DOI: 10.1128/mBio.00318-11
Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus
Abstract
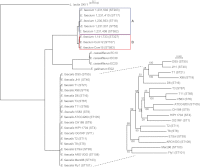
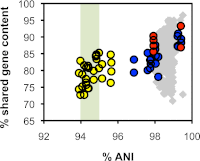
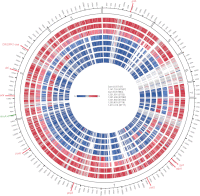
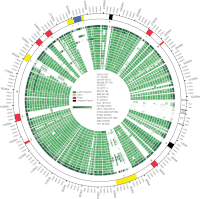
The enterococci are Gram-positive lactic acid bacteria that inhabit the gastrointestinal tracts of diverse hosts. However, Enterococcus faecium and E. faecalis have emerged as leading causes of multidrug-resistant hospital-acquired infections. The mechanism by which a well-adapted commensal evolved into a hospital pathogen is poorly understood. In this study, we examined high-quality draft genome data for evidence of key events in the evolution of the leading causes of enterococcal infections, including E. faecalis, E. faecium, E. casseliflavus, and E. gallinarum. We characterized two clades within what is currently classified as E. faecium and identified traits characteristic of each, including variation in operons for cell wall carbohydrate and putative capsule biosynthesis. We examined the extent of recombination between the two E. faecium clades and identified two strains with mosaic genomes. We determined the underlying genetics for the defining characteristics of the motile enterococci E. casseliflavus and E. gallinarum. Further, we identified species-specific traits that could be used to advance the detection of medically relevant enterococci and their identification to the species level.
Figures

References
-
- Aarestrup FM, Butaye P, Witte W. 2002. Nonhuman reservoirs of enterococci, p 55–99 In Gilmore MS, The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC
-
- Malani PN, Kauffman CA, Zervos MJ. 2002. Enterococcal disease, epidemiology, and treatment, p 385–408 In Gilmore MS, The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC
-
- Tannock GW, Cook G. 2002. Enterococci as members of the intestinal microflora of humans, p 101–132 In Gilmore MS, The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC
-
- Paulsen IT, et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
