Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing
- PMID: 22354987
- PMCID: PMC3318588
- DOI: 10.1128/MCB.06536-11
Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing
Abstract
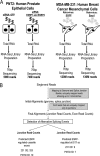
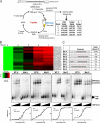
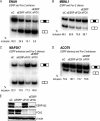
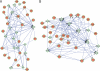
Tissue-specific alternative splicing is achieved through the coordinated assembly of RNA binding proteins at specific sites to enhance or silence splicing at nearby splice sites. We used high-throughput sequencing (RNA-Seq) to investigate the complete spectrum of alternative splicing events that are regulated by the epithelium-specific splicing regulatory proteins ESRP1 and ESRP2. We also combined this analysis with direct RNA sequencing (DRS) to reveal ESRP-mediated regulation of alternative polyadenylation. To define binding motifs that mediate direct regulation of splicing and polyadenylation by ESRP, SELEX-Seq analysis was performed, coupling traditional SELEX with high-throughput sequencing. Identification and scoring of high-affinity ESRP1 binding motifs within ESRP target genes allowed the generation of RNA maps that define the position-dependent activity of the ESRPs in regulating cassette exons and alternative 3' ends. These extensive analyses provide a comprehensive picture of the functions of the ESRPs in an epithelial posttranscriptional gene expression program.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
