A balance between Tel1 and Rif2 activities regulates nucleolytic processing and elongation at telomeres
- PMID: 22354991
- PMCID: PMC3347235
- DOI: 10.1128/MCB.06547-11
A balance between Tel1 and Rif2 activities regulates nucleolytic processing and elongation at telomeres
Abstract
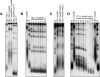
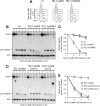
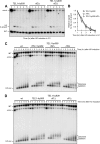
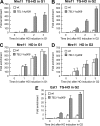
Generation of G-strand overhangs at Saccharomyces cerevisiae yeast telomeres depends primarily on the MRX (Mre11-Rad50-Xrs2) complex, which is also necessary to maintain telomere length by recruiting the Tel1 kinase. MRX physically interacts with Rif2, which inhibits both resection and elongation of telomeres. We provide evidence that regulation of telomere processing and elongation relies on a balance between Tel1 and Rif2 activities. Tel1 regulates telomere nucleolytic processing by promoting MRX activity. In fact, the lack of Tel1 impairs MRX-dependent telomere resection, which is instead enhanced by the Tel1-hy909 mutant variant, which causes telomerase-dependent telomere overelongation. The Tel1-hy909 variant is more robustly associated than wild-type Tel1 to double-strand-break (DSB) ends carrying telomeric repeat sequences. Furthermore, it increases the persistence at a DSB adjacent to telomeric repeats of both MRX and Est1, which in turn likely account for the increased telomere resection and elongation in TEL1-hy909 cells. Strikingly, Rif2 is unable to negatively regulate processing and lengthening at TEL1-hy909 telomeres, indicating that the Tel1-hy909 variant overcomes the inhibitory activity exerted by Rif2 on MRX. Altogether, these findings highlight a primary role of Tel1 in overcoming Rif2-dependent negative regulation of MRX activity in telomere resection and elongation.
Figures






References
-
- Bianchi A, Negrini S, Shore D. 2004. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell 16:139–146 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
