Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers
- PMID: 22354992
- PMCID: PMC3347240
- DOI: 10.1128/MCB.06717-11
Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers
Abstract
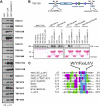
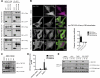
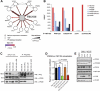
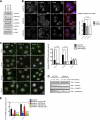
Autophagy is an evolutionarily conserved degradation pathway characterized by dynamic rearrangement of membranes that sequester cytoplasm, protein aggregates, organelles, and pathogens for delivery to the vacuole and lysosome, respectively. The ability of autophagosomal membranes to act selectively toward specific cargo is dependent on the small ubiquitin-like modifier ATG8/LC3 and the LC3-interacting region (LIR) present in autophagy receptors. Here, we describe a comprehensive protein-protein interaction analysis of TBC (Tre2, Bub2, and Cdc16) domain-containing Rab GTPase-activating proteins (GAPs) as potential autophagy adaptors. We identified 14 TBC domain-containing Rab GAPs that bind directly to ATG8 modifiers and that colocalize with LC3-positive autophagy membranes in cells. Intriguingly, one of our screening hits, TBC1D5, contains two LIR motifs. The N-terminal LIR was critical for interaction with the retromer complex and transport of cargo. Direct binding of the retromer component VPS29 to TBC1D5 could be titrated out by LC3, indicating a molecular switch between endosomes and autophagy. Moreover, TBC1D5 could bridge the endosome and autophagosome via its C-terminal LIR motif. During starvation-induced autophagy, TBC1D5 was relocalized from endosomal localization to the LC3-positive autophagosomes. We propose that LC3-interacting Rab GAPs are implicated in the reprogramming of the endocytic trafficking events under starvation-induced autophagy.
Figures

Similar articles
-
PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.Mol Cell. 2015 Jan 8;57(1):39-54. doi: 10.1016/j.molcel.2014.11.006. Epub 2014 Dec 11. Mol Cell. 2015. PMID: 25498145
-
Autophagy captures the retromer-TBC1D5 complex to inhibit receptor recycling.Autophagy. 2024 Apr;20(4):863-882. doi: 10.1080/15548627.2023.2281126. Epub 2023 Nov 17. Autophagy. 2024. PMID: 37938196 Free PMC article.
-
The LIR motif - crucial for selective autophagy.J Cell Sci. 2013 Aug 1;126(Pt 15):3237-47. doi: 10.1242/jcs.126128. J Cell Sci. 2013. PMID: 23908376 Review.
-
Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy.Elife. 2014 Feb 25;3:e01612. doi: 10.7554/eLife.01612. Elife. 2014. PMID: 24569479 Free PMC article.
-
The integration of autophagy and cellular trafficking pathways via RAB GAPs.Autophagy. 2015;11(12):2393-7. doi: 10.1080/15548627.2015.1110668. Autophagy. 2015. PMID: 26565612 Free PMC article. Review.
Cited by
-
Regulation of endoplasmic reticulum turnover by selective autophagy.Nature. 2015 Jun 18;522(7556):354-8. doi: 10.1038/nature14498. Epub 2015 Jun 3. Nature. 2015. PMID: 26040720
-
VPS35 and retromer dysfunction in Parkinson's disease.Philos Trans R Soc Lond B Biol Sci. 2024 Apr 8;379(1899):20220384. doi: 10.1098/rstb.2022.0384. Epub 2024 Feb 19. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38368930 Free PMC article. Review.
-
The Rab GTPase activating protein TBC-2 regulates endosomal localization of DAF-16 FOXO and lifespan.PLoS Genet. 2022 Aug 1;18(8):e1010328. doi: 10.1371/journal.pgen.1010328. eCollection 2022 Aug. PLoS Genet. 2022. PMID: 35913999 Free PMC article.
-
Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae.Traffic. 2015 Feb;16(2):172-90. doi: 10.1111/tra.12240. Epub 2014 Dec 16. Traffic. 2015. PMID: 25385507 Free PMC article.
-
Interplay of autophagy, receptor tyrosine kinase signalling and endocytic trafficking.Essays Biochem. 2017 Dec 12;61(6):597-607. doi: 10.1042/EBC20170091. Print 2017 Dec 12. Essays Biochem. 2017. PMID: 29233871 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
