Altered replication in human cells promotes DMPK (CTG)(n) · (CAG)(n) repeat instability
- PMID: 22354993
- PMCID: PMC3347245
- DOI: 10.1128/MCB.06727-11
Altered replication in human cells promotes DMPK (CTG)(n) · (CAG)(n) repeat instability
Abstract
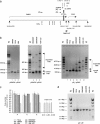
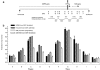
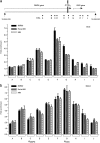
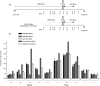
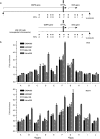
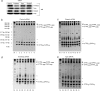
Myotonic dystrophy type 1 (DM1) is associated with expansion of (CTG)(n) · (CAG)(n) trinucleotide repeats (TNRs) in the 3' untranslated region (UTR) of the DMPK gene. Replication origins are cis-acting elements that potentiate TNR instability; therefore, we mapped replication initiation sites and prereplication complex protein binding within the ~10-kb DMPK/SIX5 locus in non-DM1 and DM1 cells. Two origins, IS(DMPK) and IS(SIX5), flanked the (CTG)(n) · (CAG)(n) TNRs in control cells and in DM1 cells. Orc2 and Mcm4 bound near each of the replication initiation sites, but a dramatic change in (CTG)(n) · (CAG)(n) replication polarity was not correlated with TNR expansion. To test whether (CTG)(n) · (CAG)(n) TNRs are cis-acting elements of instability in human cells, model cell lines were created by integration of cassettes containing the c-myc replication origin and (CTG)(n) · (CAG)(n) TNRs in HeLa cells. Replication forks were slowed by (CTG)(n) · (CAG)(n) TNRs in a length-dependent manner independent of replication polarity, implying that expanded (CTG)(n) · (CAG)(n) TNRs lead to replication stress. Consistent with this prediction, TNR instability increased in the HeLa model cells and DM1 cells upon small interfering RNA (siRNA) knockdown of the fork stabilization protein Claspin, Timeless, or Tipin. These results suggest that aberrant DNA replication and TNR instability are linked in DM1 cells.
Figures


Similar articles
-
Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells.Hum Mol Genet. 2013 Dec 20;22(25):5276-87. doi: 10.1093/hmg/ddt386. Epub 2013 Aug 9. Hum Mol Genet. 2013. PMID: 23933738 Free PMC article.
-
Oligodeoxynucleotide binding to (CTG) · (CAG) microsatellite repeats inhibits replication fork stalling, hairpin formation, and genome instability.Mol Cell Biol. 2013 Feb;33(3):571-81. doi: 10.1128/MCB.01265-12. Epub 2012 Nov 19. Mol Cell Biol. 2013. PMID: 23166299 Free PMC article.
-
Stabilization of expanded (CTG)•(CAG) repeats by antisense oligonucleotides.Mol Ther. 2011 Dec;19(12):2222-7. doi: 10.1038/mt.2011.191. Epub 2011 Oct 4. Mol Ther. 2011. PMID: 21971425 Free PMC article.
-
Myotonic dystrophy--a multigene disorder.Brain Res Bull. 2001 Oct-Nov 1;56(3-4):389-95. doi: 10.1016/s0361-9230(01)00656-6. Brain Res Bull. 2001. PMID: 11719277 Review.
-
DM1 Phenotype Variability and Triplet Repeat Instability: Challenges in the Development of New Therapies.Int J Mol Sci. 2020 Jan 10;21(2):457. doi: 10.3390/ijms21020457. Int J Mol Sci. 2020. PMID: 31936870 Free PMC article. Review.
Cited by
-
Replication fork stalling and checkpoint activation by a PKD1 locus mirror repeat polypurine-polypyrimidine (Pu-Py) tract.J Biol Chem. 2012 Sep 28;287(40):33412-23. doi: 10.1074/jbc.M112.402503. Epub 2012 Aug 6. J Biol Chem. 2012. PMID: 22872635 Free PMC article.
-
Large-scale expansions of Friedreich's ataxia GAA•TTC repeats in an experimental human system: role of DNA replication and prevention by LNA-DNA oligonucleotides and PNA oligomers.Nucleic Acids Res. 2023 Sep 8;51(16):8532-8549. doi: 10.1093/nar/gkad441. Nucleic Acids Res. 2023. PMID: 37216608 Free PMC article.
-
Dynamic DNA Energy Landscapes and Substrate Complexity in Triplet Repeat Expansion and DNA Repair.Biomolecules. 2019 Nov 6;9(11):709. doi: 10.3390/biom9110709. Biomolecules. 2019. PMID: 31698848 Free PMC article.
-
Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells.Hum Mol Genet. 2013 Dec 20;22(25):5276-87. doi: 10.1093/hmg/ddt386. Epub 2013 Aug 9. Hum Mol Genet. 2013. PMID: 23933738 Free PMC article.
-
CpG Methylation, a Parent-of-Origin Effect for Maternal-Biased Transmission of Congenital Myotonic Dystrophy.Am J Hum Genet. 2017 Mar 2;100(3):488-505. doi: 10.1016/j.ajhg.2017.01.033. Am J Hum Genet. 2017. PMID: 28257691 Free PMC article.
References
-
- Aladjem MI. 2007. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 8:588–600 - PubMed
-
- Balakumaran BS, Freudenreich CH, Zakian VA. 2000. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 9:93–100 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
