Negative feedback confers mutational robustness in yeast transcription factor regulation
- PMID: 22355134
- PMCID: PMC3309721
- DOI: 10.1073/pnas.1116360109
Negative feedback confers mutational robustness in yeast transcription factor regulation
Abstract
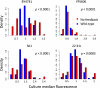
Organismal fitness depends on the ability of gene networks to function robustly in the face of environmental and genetic perturbations. Understanding the mechanisms of this stability is one of the key aims of modern systems biology. Dissecting the basis of robustness to mutation has proven a particular challenge, with most experimental models relying on artificial DNA sequence variants engineered in the laboratory. In this work, we hypothesized that negative regulatory feedback could stabilize gene expression against the disruptions that arise from natural genetic variation. We screened yeast transcription factors for feedback and used the results to establish ROX1 (Repressor of hypOXia) as a model system for the study of feedback in circuit behaviors and its impact across genetically heterogeneous populations. Mutagenesis experiments revealed the mechanism of Rox1 as a direct transcriptional repressor at its own gene, enabling a regulatory program of rapid induction during environmental change that reached a plateau of moderate steady-state expression. Additionally, in a given environmental condition, Rox1 levels varied widely across genetically distinct strains; the ROX1 feedback loop regulated this variation, in that the range of expression levels across genetic backgrounds showed greater spread in ROX1 feedback mutants than among strains with the ROX1 feedback loop intact. Our findings indicate that the ROX1 feedback circuit is tuned to respond to perturbations arising from natural genetic variation in addition to its role in induction behavior. We suggest that regulatory feedback may be an important element of the network architectures that confer mutational robustness across biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

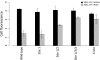


Similar articles
-
Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae.Mol Cell Biol. 1995 Nov;15(11):6109-17. doi: 10.1128/MCB.15.11.6109. Mol Cell Biol. 1995. PMID: 7565763 Free PMC article.
-
Transcript analysis of 203 novel genes from Saccharomyces cerevisiae in hap1 and rox1 mutant backgrounds.Genome. 2000 Oct;43(5):881-6. doi: 10.1139/g00-049. Genome. 2000. PMID: 11081979
-
Approaches to the study of Rox1 repression of the hypoxic genes in the yeast Saccharomyces cerevisiae.Methods. 1997 Mar;11(3):279-88. doi: 10.1006/meth.1996.0422. Methods. 1997. PMID: 9073571
-
Rox1 mediated repression. Oxygen dependent repression in yeast.Adv Exp Med Biol. 2000;475:185-95. Adv Exp Med Biol. 2000. PMID: 10849660 Review.
-
Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae.Genes (Basel). 2020 Jul 15;11(7):795. doi: 10.3390/genes11070795. Genes (Basel). 2020. PMID: 32679672 Free PMC article. Review.
Cited by
-
A multi-scale model of hepcidin promoter regulation reveals factors controlling systemic iron homeostasis.PLoS Comput Biol. 2014 Jan;10(1):e1003421. doi: 10.1371/journal.pcbi.1003421. Epub 2014 Jan 2. PLoS Comput Biol. 2014. PMID: 24391488 Free PMC article.
-
Enhancer Runaway and the Evolution of Diploid Gene Expression.PLoS Genet. 2015 Nov 12;11(11):e1005665. doi: 10.1371/journal.pgen.1005665. eCollection 2015 Nov. PLoS Genet. 2015. PMID: 26561855 Free PMC article.
-
Zinc sensing and regulation in yeast model systems.Arch Biochem Biophys. 2016 Dec 1;611:30-36. doi: 10.1016/j.abb.2016.02.031. Epub 2016 Mar 3. Arch Biochem Biophys. 2016. PMID: 26940262 Free PMC article. Review.
-
Gene Regulation and Speciation.Trends Genet. 2017 Jan;33(1):68-80. doi: 10.1016/j.tig.2016.11.003. Epub 2016 Dec 1. Trends Genet. 2017. PMID: 27914620 Free PMC article. Review.
-
Competition and evolutionary selection among core regulatory motifs in gene expression control.Nat Commun. 2023 Dec 13;14(1):8266. doi: 10.1038/s41467-023-43327-7. Nat Commun. 2023. PMID: 38092759 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

