The molten globule state is unusually deformable under mechanical force
- PMID: 22355138
- PMCID: PMC3309780
- DOI: 10.1073/pnas.1115519109
The molten globule state is unusually deformable under mechanical force
Abstract
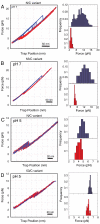
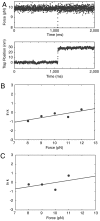
Recently, the role of force in cellular processes has become more evident, and now with advances in force spectroscopy, the response of proteins to force can be directly studied. Such studies have found that native proteins are brittle, and thus not very deformable. Here, we examine the mechanical properties of a class of intermediates referred to as the molten globule state. Using optical trap force spectroscopy, we investigated the response to force of the native and molten globule states of apomyoglobin along different pulling axes. Unlike natively folded proteins, the molten globule state of apomyoglobin is compliant (large distance to the transition state); this large compliance means that the molten globule is more deformable and the unfolding rate is more sensitive to force (the application of force or tension will have a more dramatic effect on the unfolding rate). Our studies suggest that these are general properties of molten globules and could have important implications for mechanical processes in the cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Molten globular characteristics of the native state of apomyoglobin.Nat Struct Biol. 1994 Jul;1(7):447-52. doi: 10.1038/nsb0794-447. Nat Struct Biol. 1994. PMID: 7664063
-
Thermodynamic stability of the molten globule states of apomyoglobin.J Mol Biol. 1995 Jul 7;250(2):223-38. doi: 10.1006/jmbi.1995.0373. J Mol Biol. 1995. PMID: 7608972
-
Molten globule versus variety of intermediates: influence of anions on pH-denatured apomyoglobin.FEBS Lett. 1999 Jul 23;455(3):325-31. doi: 10.1016/s0014-5793(99)00792-9. FEBS Lett. 1999. PMID: 10437798
-
Using nuclear magnetic resonance spectroscopy to study molten globule states of proteins.Methods. 2004 Sep;34(1):121-32. doi: 10.1016/j.ymeth.2004.03.009. Methods. 2004. PMID: 15283921 Review.
-
The folding process of apomyoglobin.Protein Pept Lett. 2005 Apr;12(3):229-34. doi: 10.2174/0929866053587174. Protein Pept Lett. 2005. PMID: 15777270 Review.
Cited by
-
Identification and Characterization of an Inside-Out Folding Intermediate of T4 Phage Sliding Clamp.Biophys J. 2017 Oct 17;113(8):1738-1749. doi: 10.1016/j.bpj.2017.08.043. Biophys J. 2017. PMID: 29045868 Free PMC article.
-
Catch bonds in sickle cell disease: Shear-enhanced adhesion of red blood cells to laminin.Biophys J. 2023 Jun 20;122(12):2564-2576. doi: 10.1016/j.bpj.2023.05.010. Epub 2023 May 12. Biophys J. 2023. PMID: 37177783 Free PMC article.
-
Pulling Forces Differentially Affect Refolding Pathways Due to Entangled Misfolded States in SARS-CoV-1 and SARS-CoV-2 Receptor Binding Domain.Biomolecules. 2024 Oct 18;14(10):1327. doi: 10.3390/biom14101327. Biomolecules. 2024. PMID: 39456260 Free PMC article.
-
Synthesis runs counter to directional folding of a nascent protein domain.Nat Commun. 2020 Oct 9;11(1):5096. doi: 10.1038/s41467-020-18921-8. Nat Commun. 2020. PMID: 33037221 Free PMC article.
-
Mechanochemical coupling and bi-phasic force-velocity dependence in the ultra-fast ring ATPase SpoIIIE.Elife. 2018 Mar 5;7:e32354. doi: 10.7554/eLife.32354. Elife. 2018. PMID: 29504934 Free PMC article.
References
-
- Best RB, Paci E, Hummer G, Dudko OK. Pulling direction as a reaction coordinate for the mechanical unfolding of single molecules. J Phys Chem B. 2008;112:5968–5976. - PubMed
-
- Yew ZT, Schlierf M, Rief M, Paci E. Direct evidence of the multidimensionality of the free-energy landscapes of proteins revealed by mechanical probes. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;81:031923. - PubMed
-
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

